SLFN11 Restricts LINE-1 Mobility
- PMID: 40497966
- PMCID: PMC12153781
- DOI: 10.3390/cells14110790
SLFN11 Restricts LINE-1 Mobility
Abstract
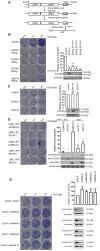
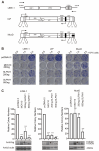
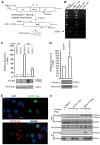
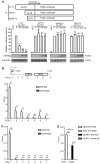
Long interspersed element-1 (LINE-1) is the only active autonomous transposon comprising about 17% of human genomes. LINE-1 transposition can cause the mutation and rearrangement of the host's genomic DNA. The host has, therefore, developed multiple mechanisms to restrict LINE-1 mobility. Here, we report that SLFN11, a member of the Schlafen family, can restrict LINE-1 retrotransposition, and the inhibitory activity requires its helicase domain. Mechanistically, SLFN11 specifically binds to the LINE-1 5' untranslated region (5'UTR) and blocks RNA polymerase II recruitment, thereby suppressing its transcription. Furthermore, SLFN11 promotes heterochromatinization, suggesting an epigenetic inhibition pathway.
Keywords: LINE-1; RNA polymerase II; SLFN11; epigenetics; helicase; heterochromatin; long interspersed element-1; transposon.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study.
Figures

Similar articles
-
The MOV10 helicase inhibits LINE-1 mobility.J Biol Chem. 2013 Jul 19;288(29):21148-21160. doi: 10.1074/jbc.M113.465856. Epub 2013 Jun 10. J Biol Chem. 2013. PMID: 23754279 Free PMC article.
-
Efficient translation initiation directed by the 900-nucleotide-long and GC-rich 5' untranslated region of the human retrotransposon LINE-1 mRNA is strictly cap dependent rather than internal ribosome entry site mediated.Mol Cell Biol. 2007 Jul;27(13):4685-97. doi: 10.1128/MCB.02138-06. Epub 2007 Apr 30. Mol Cell Biol. 2007. PMID: 17470553 Free PMC article.
-
Hepatitis B virus polymerase restricts LINE-1 mobility.Gene. 2023 Jan 20;850:146943. doi: 10.1016/j.gene.2022.146943. Epub 2022 Oct 2. Gene. 2023. PMID: 36198378
-
LINE-1 retrotransposition and its deregulation in cancers: implications for therapeutic opportunities.Genes Dev. 2023 Dec 26;37(21-24):948-967. doi: 10.1101/gad.351051.123. Genes Dev. 2023. PMID: 38092519 Free PMC article. Review.
-
[The connection between LINE-1 retrotransposition and human tumorigenesis].Yi Chuan. 2016 Feb;38(2):93-102. doi: 10.16288/j.yczz.15-470. Yi Chuan. 2016. PMID: 26907772 Review. Chinese.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

