Insulin-Degrading Enzyme Regulates mRNA Processing and May Interact with the CCR4-NOT Complex
- PMID: 40497968
- PMCID: PMC12153694
- DOI: 10.3390/cells14110792
Insulin-Degrading Enzyme Regulates mRNA Processing and May Interact with the CCR4-NOT Complex
Abstract
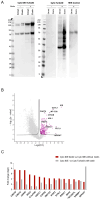
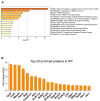
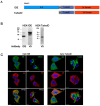
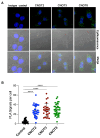
Insulin-degrading enzyme is a zinc metalloprotease that degrades low-molecular-weight substrates, including insulin. Ubiquitous expression, high evolutionary conservation, upregulation of Ide in stress situations, and literature findings suggest a broader function of Ide in cell physiology and protein homeostasis that remains to be elucidated. We used proteomics and transcriptomics approaches to search for leads related to a broader role of Ide in protein homeostasis. We combined an analysis of the proteome and single-cell transcriptome of Ide+/+ and Ide-/- pancreatic islet cells with an examination of the interactome of human cytosolic Ide using proximity biotinylation. We observe an upregulation of pathways related to RNA processing, translation and splicing in Ide+/+ relative to Ide-/- islet cells. Corroborating these results and providing a potential mechanistic explanation, proximity biotinylation reveals interaction of Ide with several subunits of CCR4-NOT, a key mRNA deadenylase regulating gene expression "from birth to death". We propose a speculative model in which human and murine Ide cooperate with CCR4-NOT to control protein expression in proteotoxic and metabolic stress situations through cooperation between their deadenylase and protease functions.
Keywords: CCR4-NOT; RNA processing; beta cell; insulinase; islet of Langerhans; protein homeostasis.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


References
-
- Zhu S., Waeckel-Énée E., Oshima M., Moser A., Bessard M.-A., Gdoura A., Roger K., Mode N., Lipecka J., Yilmaz A., et al. Islet cell stress induced by insulin-degrading enzyme deficiency promotes regeneration and protection from autoimmune diabetes. iScience. 2024;27:109929. doi: 10.1016/j.isci.2024.109929. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

