Functional Characterization of LTR12C as Regulators of Germ-Cell-Associated TA-p63 in U87-MG and T98-G In Vitro Models
- PMID: 40498028
- PMCID: PMC12154421
- DOI: 10.3390/cells14110852
Functional Characterization of LTR12C as Regulators of Germ-Cell-Associated TA-p63 in U87-MG and T98-G In Vitro Models
Abstract
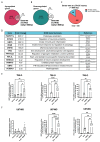
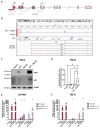
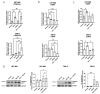
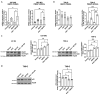
Glioblastoma multiforme (GBM) is a deadly disease known for its genetic heterogeneity. LTR12C is an endogenous retrovirus-derived regulator of pro-apoptotic genes and is normally silenced by epigenetic regulation. In this study, we found that the treatment of two glioblastoma cell lines, T98-G and U87-MG, with DNA methyltransferase (DNMT) and histone deacetylase (HDAC) inhibitors activated LTR12C expression. Combined treatment with these epigenetic drugs exerted a synergistic action on the LTR12C activation in both cell lines, while treatment with each drug as a single agent had a far weaker effect. A strong induction of the expression of the TP63 gene was seen in both cell lines, with the pro-apoptotic isoform GTA-p63 accounting for most of this increase. Coherently, downstream targets of p63, such as p21 and PUMA, were also induced by the combined treatment. Furthermore, we observed a significant reduction in the GBM cell growth and viability following the dual DNMT/HDAC inhibition. These findings reveal that the reactivation of LTR12C expression has the potential to modulate survival pathways in glioblastoma and provide information regarding possible epigenetic mechanisms that can be used to treat this deadly disease.
Keywords: GTA-p63; LTR12C; epigenetics; glioblastoma; p21.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Goyal A., Bauer J., Hey J., Papageorgiou D.N., Stepanova E., Daskalakis M., Scheid J., Dubbelaar M., Klimovich B., Schwarz D., et al. DNMT and HDAC Inhibition Induces Immunogenic Neoantigens from Human Endogenous Retroviral Element-Derived Transcripts. Nat. Commun. 2023;14:6731. doi: 10.1038/s41467-023-42417-w. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

