Using repeated lysis steps fractionates between heterotrophic and cyanobacterial DNA extracted from xenic cyanobacterial cultures
- PMID: 40498592
- PMCID: PMC12341861
- DOI: 10.1093/g3journal/jkaf135
Using repeated lysis steps fractionates between heterotrophic and cyanobacterial DNA extracted from xenic cyanobacterial cultures
Abstract
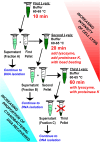
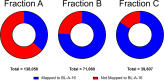
Extracting DNA from cyanobacteria can be a challenge because of their diverse morphologies, challenging cellular structure, and the heterotrophic microbiome often present within cyanobacterial cultures. As such, even with high DNA yields, the percentage of reads coming from the cyanobacterial host can be low, leading to an incomplete cyanobacterial genome assembly. In this research, we optimized a DNA isolation protocol using three iterative cell lysis steps to enrich the portion of DNA isolated coming from the cyanobacterial host rather than the heterotrophic microbiome. In order to utilize in-house nanopore sequencing, we faced a challenge using our lysis protocol: the iterative lysis approach led to more DNA shearing than is ideal for this sequencing technology. To solve this, we used two bead-based size selection steps to remove shorter molecules of DNA before nanopore sequencing. Analysis of the sequenced reads showed that, in the first lysis, the cyanobacterial sequences were only 35% of all reads. In the repeated lysis steps, however, the proportion of reads coming from the cyanobacterium increased to 75% or higher. Using our iterative lysis protocol, we were able to sequence the genomes of two fresh water cyanobacteria isolated from northern Mississippi, namely Leptolyngbya sp. BL-A-14 and Limnothrix sp. BL-A-16. The genomes of these isolates were assembled as closed chromosomes of 7.2 and 4.5 Mb for BL-A-14 and BL-A-16, respectively. As it is not always possible to prepare axenic cultures of cyanobacteria, we hope our approach will be useful for sequencing other xenic cultures of cyanobacteria.
Keywords: cyanobacteria; microbiomes; whole genome sequencing; xenic cultures.
© The Author(s) 2025. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest: The author(s) declare no conflicts of interest.
Figures

Update of
-
Using Repeated Lysis Steps Fractionates Between Heterotrophic and Cyanobacterial DNA Extracted from Xenic Cyanobacterial Cultures.bioRxiv [Preprint]. 2024 Aug 22:2024.08.22.609136. doi: 10.1101/2024.08.22.609136. bioRxiv. 2024. Update in: G3 (Bethesda). 2025 Aug 6;15(8):jkaf135. doi: 10.1093/g3journal/jkaf135. PMID: 39229059 Free PMC article. Updated. Preprint.
Similar articles
-
Using Repeated Lysis Steps Fractionates Between Heterotrophic and Cyanobacterial DNA Extracted from Xenic Cyanobacterial Cultures.bioRxiv [Preprint]. 2024 Aug 22:2024.08.22.609136. doi: 10.1101/2024.08.22.609136. bioRxiv. 2024. Update in: G3 (Bethesda). 2025 Aug 6;15(8):jkaf135. doi: 10.1093/g3journal/jkaf135. PMID: 39229059 Free PMC article. Updated. Preprint.
-
Decontamination of DNA sequences from a Streptomyces genome for optimal genome mining.Braz J Microbiol. 2025 Mar;56(1):79-89. doi: 10.1007/s42770-024-01598-2. Epub 2025 Jan 15. Braz J Microbiol. 2025. PMID: 39812972
-
Can a Liquid Biopsy Detect Circulating Tumor DNA With Low-passage Whole-genome Sequencing in Patients With a Sarcoma? A Pilot Evaluation.Clin Orthop Relat Res. 2025 Jan 1;483(1):39-48. doi: 10.1097/CORR.0000000000003161. Epub 2024 Jun 21. Clin Orthop Relat Res. 2025. PMID: 38905450
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Survivor, family and professional experiences of psychosocial interventions for sexual abuse and violence: a qualitative evidence synthesis.Cochrane Database Syst Rev. 2022 Oct 4;10(10):CD013648. doi: 10.1002/14651858.CD013648.pub2. Cochrane Database Syst Rev. 2022. PMID: 36194890 Free PMC article.
References
-
- Australian Centre for Ecogenomics . 2024. Genome Taxonomy Database. Release 09-RS220. [accessed 2024 Sep 13]. https://gtdb.ecogenomic.org/.
-
- Berdalet E, Fleming LE, Gowen R, Davidson K, Hess P, Backer LC, Moore SK, Hoagland P, Enevoldsen H. 2016. Marine harmful algal blooms, human health and wellbeing: challenges and opportunities in the 21st century. J Mar Biol Assoc U K. 96(1):61–91. doi: 10.1017/S0025315415001733. - DOI - PMC - PubMed
-
- Blin K, Shaw S, Augustijn HE, Reitz ZL, Biermann F, Alanjary M, Fetter A, Terlouw BR, Metcalf WW, Helfrich EJN, et al. 2023. antiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 51(W1):W46–W50. doi: 10.1093/nar/gkad344. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
