Molecular patterns and mechanisms of tumorigenesis in HPV-associated and HPV-independent sinonasal squamous cell carcinoma
- PMID: 40500270
- PMCID: PMC12159145
- DOI: 10.1038/s41467-025-59409-7
Molecular patterns and mechanisms of tumorigenesis in HPV-associated and HPV-independent sinonasal squamous cell carcinoma
Abstract
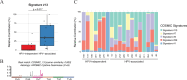
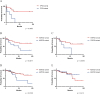
Mechanisms of tumorigenesis in sinonasal squamous cell carcinoma (SNSCC) remain poorly understood due to its rarity. A subset of SNSCC is associated with human papillomavirus (HPV), but it is unclear whether HPV drives tumorigenesis or acts as a neutral bystander. Here, we show that HPV-associated SNSCC shares mutational patterns found in HPV-associated cervical and head and neck squamous cell carcinoma, including lack of TP53 mutations, hotspot mutations in PI3K and FGFR3, enrichment of APOBEC mutagenesis, viral integration at known hotspots, and frequent epigenetic regulator alterations. We identify HPV-associated SNSCC-specific recurrent mutations in KMT2C, UBXN11, AP3S1, MT-ND4, and MT-ND5, with KMT2D and FGFR3 mutations correlating with reduced overall survival. We establish an HPV-associated SNSCC cell line, showing that combinatorial small-molecule inhibition of YAP/TAZ and PI3K synergistically suppresses clonogenicity. Combining YAP/TAZ blockade with vertical PI3K inhibition may benefit HPV-associated SNSCC, whereas targeting MYC and horizontal inhibition of RAS/PI3K may suit HPV-independent SNSCC.
© 2025. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Competing interests: This research was supported (in part) by the Intramural Research Program of the NIH, Center for Cancer Research, National Cancer Institute. This study was supported in part by a research grant from Investigator-Initiated Studies Program of Merck Sharp & Dohme, LLC (N. London). The opinions expressed in this paper are those of the authors and do not necessarily represent those of Merck Sharp & Dohme, LLC. All other authors declare no competing interests.
Figures





Update of
-
Molecular patterns and mechanisms of tumorigenesis in HPV-associated and HPV-independent sinonasal squamous cell carcinoma.bioRxiv [Preprint]. 2024 Jun 18:2024.06.17.598514. doi: 10.1101/2024.06.17.598514. bioRxiv. 2024. Update in: Nat Commun. 2025 Jun 11;16(1):5285. doi: 10.1038/s41467-025-59409-7. PMID: 38979305 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

