Overcoming intra-tumoral heterogeneity for biomarker discovery in the high-grade serous ovarian cancer proteome
- PMID: 40500315
- PMCID: PMC12159146
- DOI: 10.1038/s41698-025-00911-y
Overcoming intra-tumoral heterogeneity for biomarker discovery in the high-grade serous ovarian cancer proteome
Abstract
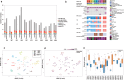
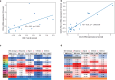
Improved biomarkers of treatment response are needed for patients with high-grade serous ovarian cancer (HGSC). A challenge is substantial anatomical site-to-site variation in expression. We completed data-independent acquisition-mass spectrometry (DIA-MS) analysis of 404 fresh frozen and 78 formalin-fixed, paraffin-embedded HGSC tissue samples from the ovary (adnexal) and a common secondary site (omentum) in 11 patients. This was compared with mutation testing, gene expression, and whole-genome copy number profiling. Proteins with relatively stable intra- and variable inter-individual expression (n = 1651), included a 52-protein module reflecting interferon-mediated tissue inflammation, indicative of a cGAS-STING pathway cytosolic double-stranded (ds) DNA response. The dsDNA sensing/inflammation score was higher in the omentum compared with the ovary. Ovarian HGSC samples showed marked inter-individual differences in inflammatory and immune responses to DNA damage. Stable discriminative features of the HGSC proteome, a prerequisite for clinical predictive biomarkers, are detectable in ovary (adnexal) tissue samples.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: A.DeF. and R.L.B. have received research support from Illumina for unrelated work. The remaining authors declare no competing interests.
Figures



References
-
- Lorusso, D. et al. Newly diagnosed ovarian cancer: which first-line treatment?. Cancer Treat. Rev.91, 102111 (2020). - PubMed
-
- Moore, K. et al. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N. Engl. J. Med.379, 2495–2505 (2018). - PubMed
-
- Patch, A. M. et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature521, 489–494 (2015). - PubMed
-
- Zhang, A. W. et al. Interfaces of malignant and immunologic clonal dynamics in ovarian cancer. Cell173, 1755–1769 e1722 (2018). - PubMed
Grants and funding
- 2017/ECF004/Cancer Institute NSW Early Career Fellowship
- Grants 1186505 and 2029088/National Health and Medical Research Council of Australia
- ID400413, ID400281/National Health and Medical Research Council of Australia
- ID400413, ID400281/National Health and Medical Research Council of Australia
- Award No. W81XWH-21-1-0401/U.S. Army Medical Research and Materiel Command Ovarian Cancer Research Program
- MCRF22018/Victorian Cancer Agency
- DAMD17-01-1-0729/U.S. Army Medical Research and Materiel Command
- IG 18-01/Cancer Council NSW
- IG 18-01/Cancer Council NSW
- IG 18-01/Cancer Council NSW
- IG 18-01/Cancer Council NSW
- IG 18-01/Cancer Council NSW
- 2017/TPG001, REG171150/Cancer Institute NSW
- 2017/TPG001, REG171150/Cancer Institute NSW
- 2017/TPG001, REG171150/Cancer Institute NSW
- 2017/TPG001, REG171150/Cancer Institute NSW
- CMP-01/NSW Ministry of Health
- CMP-01/NSW Ministry of Health
- MRFF-PD/Medical Research Futures Fund
- MRFF-PD/Medical Research Futures Fund
- GNT1170739/National Health and Medical Research Council (NHMRC) of Australia European Union grant
- GNT1170739/National Health and Medical Research Council (NHMRC) of Australia European Union grant
- GNT1170739/National Health and Medical Research Council (NHMRC) of Australia European Union grant
- GNT1170739/National Health and Medical Research Council (NHMRC) of Australia European Union grant
- GNT1137064/NHMRC Fellowship
LinkOut - more resources
Full Text Sources
Research Materials

