T and B cell responses against Epstein-Barr virus in primary sclerosing cholangitis
- PMID: 40500415
- PMCID: PMC12283410
- DOI: 10.1038/s41591-025-03692-w
T and B cell responses against Epstein-Barr virus in primary sclerosing cholangitis
Abstract
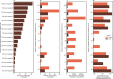
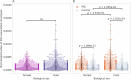
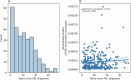
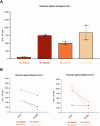
Primary sclerosing cholangitis (PSC) is an idiopathic, progressive and incurable liver disease. Here, we aimed for systematic analyses of adaptive immune responses in PSC. By profiling the T cell repertoires of 504 individuals with PSC and 904 healthy controls, we identified 1,008 clonotypes associated with PSC. A substantial fraction of these clonotypes was restricted to known PSC human leukocyte antigen susceptibility alleles and known to target Epstein-Barr virus (EBV) epitopes. We further utilized phage-immunoprecipitation sequencing to determine antibody epitope repertoires of 120 individuals with PSC and 202 healthy controls, which showed a higher burden of anti-EBV responses in PSC than controls. EBV-specific monoclonal antibodies isolated from B cells in PSC livers corroborated convergent B and T cell responses against EBV. By analyzing electronic health records of >116 million people, we identified an association between infectious mononucleosis and PSC (odds ratio, 12; 95% confidence interval, 6.3-22.9), suggesting a link between EBV and PSC.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: M.P., D.H.M., B.H. and H.S.R. have or had employment and equity ownership with Adaptive Biotechnologies. H.E. did an internship at Adaptive biotechnologies from July 2023 until September 2023. F.T. received speaker’s fees from Abbvie, Bristol-Myers-Squibb, Celltrion Healthcare, Dr Falk Pharma, Eli Lilly, Ferring Pharmaceuticals and Janssen, and funding from Sanofi/Regeneron. T.H.K. declares consulting fees from Albireo, Boehringer Ingelheim, MSD, Gilead and Falk Pharma unrelated to the present work. A.-K.P. (institution) has received speaker, consulting or travel honoraria from Novartis, Biogen, Roche and UCB all used for research support and unrelated to the present work. The other authors declare no competing interests.
Figures




References
-
- Karlsen, T. H., Folseraas, T., Thorburn, D. & Vesterhus, M. Primary sclerosing cholangitis; a comprehensive review. J. Hepatol.67, 1298–1323 (2017). - PubMed
-
- Chazouilleres, O. et al. EASL clinical practice guidelines on sclerosing cholangitis. J. Hepatol.77, 761–806 (2022). - PubMed
-
- van Munster, K. N. et al. Disease burden in primary sclerosing cholangitis in the Netherlands: a long-term follow-up study. Liver Int.43, 639–648 (2023). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

