A map of blood regulatory variation in South Africans enables GWAS interpretation
- PMID: 40500424
- PMCID: PMC12283414
- DOI: 10.1038/s41588-025-02223-0
A map of blood regulatory variation in South Africans enables GWAS interpretation
Abstract
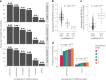
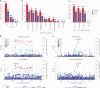
Functional genomics resources are critical for interpreting human genetic studies, but currently they are predominantly from European-ancestry individuals. Here we present the South African Blood Regulatory (SABR) resource, a map of blood regulatory variation that includes three South Eastern Bantu-speaking groups. Using paired whole-genome and blood transcriptome data from over 600 individuals, we map the genetic architecture of 40 blood cell traits derived from deconvolution analysis, as well as expression, splice and cell-type interaction quantitative trait loci. We comprehensively compare SABR to the Genotype Tissue Expression Project and characterize thousands of regulatory variants only observed in African-ancestry individuals. Finally, we demonstrate the increased utility of SABR for interpreting African-ancestry association studies by identifying putatively causal genes and molecular mechanisms through colocalization analysis of blood-relevant traits from the Pan-UK Biobank. Importantly, we make full SABR summary statistics publicly available to support the African genomics community.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: S.E.C., A.K.-E., M.K., O.A.G., M.H., S.L.v.B., E.E.B., S.K., K.-D.H.N., K.A.W. and L.Y.-A. were or are employees and/or equity owners at Variant Bio. All other authors declare no competing interests.
Figures



References
-
- Uffelmann, E. et al. Genome-wide association studies. Nat. Rev. Methods Primers1, 59 (2021).
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

