Targeting GRPR for sex hormone-dependent cancer after loss of E-cadherin
- PMID: 40500450
- PMCID: PMC12267067
- DOI: 10.1038/s41586-025-09111-x
Targeting GRPR for sex hormone-dependent cancer after loss of E-cadherin
Erratum in
-
Publisher Correction: Targeting GRPR for sex hormone-dependent cancer after loss of E-cadherin.Nature. 2025 Jul;643(8074):E28. doi: 10.1038/s41586-025-09353-9. Nature. 2025. PMID: 40659787 Free PMC article. No abstract available.
Abstract
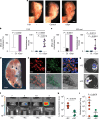
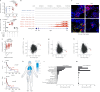
Sex inequalities in cancer are well documented, but the current limited understanding is hindering advances in precision medicine and therapies1. Consideration of ethnicity, age and sex is essential for the management of cancer patients because they underlie important differences in both incidence and response to treatment2,3. Age-related hormone production, which is a consistent divergence between the sexes, is underestimated in cancers that are not recognized as being hormone dependent4-6. Here, we show that premenopausal women have increased vulnerability to cancers, and we identify the cell-cell adhesion molecule E-cadherin as a crucial component in the oestrogen response in various cancers, including melanoma. In a mouse model of melanoma, we discovered an oestrogen-sensitizing pathway connecting E-cadherin, β-catenin, oestrogen receptor-α and GRPR that promotes melanoma aggressiveness in women. Inhibiting this pathway by targeting GRPR or oestrogen receptor-α reduces metastasis in mice, indicating its therapeutic potential. Our study introduces a concept linking hormone sensitivity and tumour phenotype in which hormones affect cell phenotype and aggressiveness. We have identified an integrated pro-tumour pathway in women and propose that targeting a G-protein-coupled receptor with drugs not commonly used for cancer treatment could be more effective in treating E-cadherin-dependent cancers in women. This study emphasizes the importance of sex-specific factors in cancer management and offers hope of improving outcomes in various cancers.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures













References
-
- Dong, M. et al. Sex differences in cancer incidence and survival: a pan-cancer analysis. Cancer Epidemiol. Biomarkers Prev.29, 1389–1397 (2020). - PubMed
-
- Zhao, Y., Wang, X., Liu, Y., Wang, H.-Y. & Xiang, J. The effects of estrogen on targeted cancer therapy drugs. Pharmacol. Res.177, 106131 (2022). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

