Metabolic adaptations direct cell fate during tissue regeneration
- PMID: 40500453
- PMCID: PMC12240837
- DOI: 10.1038/s41586-025-09097-6
Metabolic adaptations direct cell fate during tissue regeneration
Erratum in
-
Publisher Correction: Metabolic adaptations direct cell fate during tissue regeneration.Nature. 2025 Jul;643(8072):E15. doi: 10.1038/s41586-025-09294-3. Nature. 2025. PMID: 40595366 Free PMC article. No abstract available.
Abstract
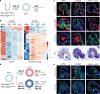
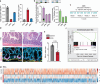
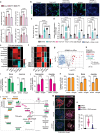
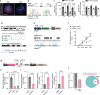
Although cell-fate specification is generally attributed to transcriptional regulation, emerging data also indicate a role for molecules linked with intermediary metabolism. For example, α-ketoglutarate (αKG), which fuels energy production and biosynthetic pathways in the tricarboxylic acid (TCA) cycle, is also a co-factor for chromatin-modifying enzymes1-3. Nevertheless, whether TCA-cycle metabolites regulate cell fate during tissue homeostasis and regeneration remains unclear. Here we show that TCA-cycle enzymes are expressed in the intestine in a heterogeneous manner, with components of the αKG dehydrogenase complex4-6 upregulated in the absorptive lineage and downregulated in the secretory lineage. Using genetically modified mouse models and organoids, we reveal that 2-oxoglutarate dehydrogenase (OGDH), the enzymatic subunit of the αKG dehydrogenase complex, has a dual, lineage-specific role. In the absorptive lineage, OGDH is upregulated by HNF4 transcription factors to maintain the bioenergetic and biosynthetic needs of enterocytes. In the secretory lineage, OGDH is downregulated through a process that, when modelled, increases the levels of αKG and stimulates the differentiation of secretory cells. Consistent with this, in mouse models of colitis with impaired differentiation and maturation of secretory cells, inhibition of OGDH or supplementation with αKG reversed these impairments and promoted tissue healing. Hence, OGDH dependency is lineage-specific, and its regulation helps to direct cell fate, offering insights for targeted therapies in regenerative medicine.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: A patent application (PCT/US2024/027206) has been published as WO2024229094A1 ( https://patents.google.com/patent/WO2024229094A1/en?oq=PCT%2fUS2024%2f027206 ; ref. 50). The patent covers methods for treating gastrointestinal inflammatory diseases using αKG modulators. A.C.-P. and S.W.L. are listed as inventors. S.W.L. is a founder and member of the scientific advisory board of Blueprint Medicines, Mirimus, ORIC Pharmaceuticals, Senescea, and Faeth Therapeutics, is on the scientific advisory board of PMV Pharmaceuticals and is a consultant for Fate Therapeutics. The remaining authors declare no competing interests.
Figures










References
-
- Armstrong, C. T., Anderson, J. L. & Denton, R. M. Studies on the regulation of the human E1 subunit of the 2-oxoglutarate dehydrogenase complex, including the identification of a novel calcium-binding site. Biochem. J.459, 369–381 (2014). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

