In vivo transition in chromatin accessibility during differentiation of deep-layer excitatory neurons in the neocortex
- PMID: 40501413
- PMCID: PMC12268177
- DOI: 10.1242/dev.204564
In vivo transition in chromatin accessibility during differentiation of deep-layer excitatory neurons in the neocortex
Abstract
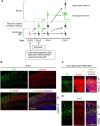
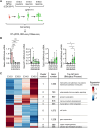
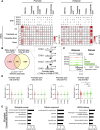
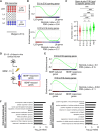
During neuronal differentiation, gene transcription patterns change in response to both intrinsic and extrinsic cues. Chromatin regulation at regulatory elements plays a key role in this process. However, how chromatin accessibility evolves in vivo in cortical neurons remains unclear. Here, we established a method for labeling differentiating neurons with specific birthdates. Using this method, we traced the 4-day differentiation process of in vivo deep-layer excitatory neurons in the mouse embryonic cortex and examined changes in the genome-wide transcription pattern and chromatin accessibility using RNA sequencing and DNase sequencing, respectively. We found that genomic regions of genes linked to mature neuronal functions, including deep layer-specific and stimulus-responsive genes, became accessible even at the embryonic stage. Additionally, our results indicated the involvement of bivalent marks in neural precursor/stem cells and Dmrt3 and Dmrta2 in the regulation of chromatin accessibility during neuronal differentiation. These findings highlight the importance of chromatin regulation in embryonic neurons, enabling the timely activation of neuronal genes during maturation.
Keywords: Bivalent genes; Chromatin accessibility; Cortical neurons; Differentiation; Dmrt; Mouse; Transcriptome.
© 2025. Published by The Company of Biologists.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures


Similar articles
-
Transcription Factor EB Overexpression through Glial Fibrillary Acidic Protein Promoter Disrupts Neuronal Lamination by Dysregulating Neurogenesis during Embryonic Development.Dev Neurosci. 2025;47(1):40-54. doi: 10.1159/000538656. Epub 2024 Apr 18. Dev Neurosci. 2025. PMID: 38583418 Free PMC article.
-
The chromatin remodeler ADNP regulates neurodevelopmental disorder risk genes and neocortical neurogenesis.Proc Natl Acad Sci U S A. 2025 Jan 21;122(3):e2405981122. doi: 10.1073/pnas.2405981122. Epub 2025 Jan 14. Proc Natl Acad Sci U S A. 2025. PMID: 39808658 Free PMC article.
-
Epigenetic modifiers promote mitochondrial biogenesis and oxidative metabolism leading to enhanced differentiation of neuroprogenitor cells.Cell Death Dis. 2018 Mar 2;9(3):360. doi: 10.1038/s41419-018-0396-1. Cell Death Dis. 2018. PMID: 29500414 Free PMC article.
-
Stage-specific DNA methylation dynamics in mammalian heart development.Epigenomics. 2025 Apr;17(5):359-371. doi: 10.1080/17501911.2025.2467024. Epub 2025 Feb 21. Epigenomics. 2025. PMID: 39980349 Review.
-
Polycomb function in early mouse development.Cell Death Differ. 2025 Jan;32(1):90-99. doi: 10.1038/s41418-024-01340-3. Epub 2024 Jul 12. Cell Death Differ. 2025. PMID: 38997437 Review.
References
-
- Arnold, S. J., Huang, G.-J., Cheung, A. F. P., Era, T., Nishikawa, S.-I., Bikoff, E. K., Molnár, Z., Robertson, E. J. and Groszer, M. (2008). The T-box transcription factor Eomes/Tbr2 regulates neurogenesis in the cortical subventricular zone. Genes Dev. 22, 2479-2484. 10.1101/gad.475408 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 16H06279, JP22H04687, 23H04214, and 24H01227/Japan Society for the Promotion of Science
- 22H00431/Japan Society for the Promotion of Science
- 23gm1310004/Japan Society for the Promotion of Science
- 22gm6110021/Japan Society for the Promotion of Science
- Takeda Science Foundation
- Uehara Memorial Foundation
- Asahi Glass Foundation
- Chugai Foundation for Innovative Drug Discovery Science
- Astellas Foundation for Research on Metabolic Disorders
- Naito Foundation
- Secom Science and Technology Foundation
- Ono Pharmaceutical Foundation for Oncology, Immunology, and Neurology
- Kurata Memorial Hitachi Science and Technology Foundation
- JP23gm1310004/Japan Agency for Medical Research and Development
- JP22gm6110021/Japan Agency for Medical Research and Development
- Ministry of Education, Culture, Sports, Science and Technology
- JP22H00431/Japan Society for the Promotion of Science
- 24H01227/Japan Society for the Promotion of Science
- 23H04214/Japan Society for the Promotion of Science
- JP22H04687/Japan Society for the Promotion of Science
- 16H06279/Japan Society for the Promotion of Science
- SECOM Science and Technology Foundation
- Ono Pharmaceutical
- Hitachi Global Foundation
- University of Tokyo
LinkOut - more resources
Full Text Sources
Research Materials

