This is a preprint.
Categorizing prediction modes within low-pLDDT regions of AlphaFold2 structures
- PMID: 40501571
- PMCID: PMC12157579
- DOI: 10.1101/2025.06.06.658382
Categorizing prediction modes within low-pLDDT regions of AlphaFold2 structures
Update in
-
Categorizing prediction modes within low-pLDDT regions of AlphaFold2 structures: near-predictive, pseudostructure and barbed wire.Acta Crystallogr D Struct Biol. 2025 Oct 1;81(Pt 10):558-572. doi: 10.1107/S2059798325007843. Epub 2025 Sep 12. Acta Crystallogr D Struct Biol. 2025. PMID: 40937679 Free PMC article.
Abstract
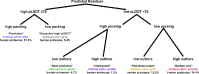
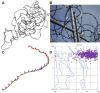
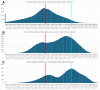
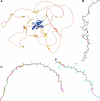
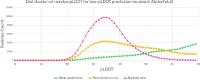
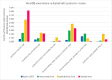
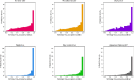
AlphaFold2 protein structure predictions are widely available for structural biology uses. These predictions, especially for eukaryotic proteins, frequently contain extensive regions predicted below the pLDDT 70 level, the rule-of-thumb cutoff for high confidence. This work identifies major modes of behavior within low-pLDDT regions through a survey of human proteome predictions provided by the AlphaFold Protein Structure Database. The near-predictive mode resembles folded protein and can be a nearly accurate prediction. Barbed wire is extremely unproteinlike, being recognized by wide looping coils, an absence of packing contacts, and numerous signature validation outliers, and it likely represents a nonpredicted region. Pseudostructure presents an intermediate behavior with a misleading appearance of isolated and badly formed secondary structure-like elements. These prediction modes are compared with annotations of disorder from MobiDB, showing general correlation between barbed wire/pseudostructure and many measures of disorder, an association between pseudostructure and signal peptides, and an association between near-predictive and regions of conditional folding. To enable users to identify these regions within a prediction, a new Phenix tool is developed encompassing the results of this work, including prediction annotation, visual markup, and residue selection based on these prediction modes. This tool will help users develop expertise in interpreting difficult AlphaFold predictions and identify the near-predictive regions that can aid in molecular replacement when a prediction does not contain enough high-pLDDT regions.
Keywords: AlphaFold; barbed wire; conditional folding; low pLDDT; near-predictive; signal peptides; structure prediction; structure validation.
Conflict of interest statement
Conflicts of interest The authors declare no conflicts of interest.
Figures
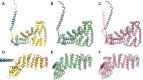
References
-
- Abramson J., Adler J., Dunger J., Evans R., Green T., Pritzel A., Ronneberger O., Willmore L., Ballard A. J., Bambrick J., Bodenstein S. W., Evans D. A., Hung C. C., O’Neill M., Reiman D., Tunyasuvunakool K., Wu Z., Zemgulyte A., Arvaniti E., Beattie C., Bertolli O., Bridgland A., Cherepanov A., Congreve M., Cowen-Rivers A. I., Cowie A., Figurnov M., Fuchs F. B., Gladman H., Jain R., Khan Y. A., Low C. M. R., Perlin K., Potapenko A., Savy P., Singh S., Stecula A., Thillaisundaram A., Tong C., Yakneen S., Zhong E. D., Zielinski M., Zidek A., Bapst V., Kohli P., Jaderberg M., Hassabis D. & Jumper J. M. (2024). Nature 630, 493–500. - PMC - PubMed
-
- Adzhubei A. A., Sternberg M. J. & Makarov A. A. (2013). J Mol Biol 425, 2100–2132. - PubMed
-
- Baek M., DiMaio F., Anishchenko I., Dauparas J., Ovchinnikov S., Lee G. R., Wang J., Cong Q., Kinch L. N., Schaeffer R. D., Millan C., Park H., Adams C., Glassman C. R., DeGiovanni A., Pereira J. H., Rodrigues A. V., van Dijk A. A., Ebrecht A. C., Opperman D. J., Sagmeister T., Buhlheller C., Pavkov-Keller T., Rathinaswamy M. K., Dalwadi U., Yip C. K., Burke J. E., Garcia K. C., Grishin N. V., Adams P. D., Read R. J. & Baker D. (2021). Science 373, 871–876. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
