This is a preprint.
eIF3d and eIF3e mediate selective translational control of hypoxia that can be inhibited by novel small molecules
- PMID: 40501575
- PMCID: PMC12154685
- DOI: 10.1101/2025.05.29.656739
eIF3d and eIF3e mediate selective translational control of hypoxia that can be inhibited by novel small molecules
Abstract
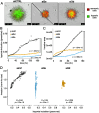
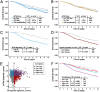
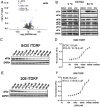
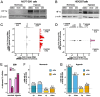
Exposure to hypoxia is linked to increased cellular plasticity and enhanced metastasis; effects which are primarily attributed to the transcriptional activation of large gene programs downstream of hypoxia inducible factors (HIFs). However, translational effects in hypoxia, that likely precede transcriptional effects, have remained largely unexplored. Using ribosome-profiling, we uncovered a selective translational response in acute hypoxia that is eIF3d/eIF3e-dependent and controls downstream hypoxic responses including HIF1a accumulation and cellular invasion. We further demonstrated that eIF3e copy number and an eIF3e-expression signature are associated with worsened outcomes for breast cancer patients. Finally, we identified a class of novel small molecules that target eIF3e specifically, reducing the translational response to hypoxia and to ER stress, another stressor that is dependent on eIF3d/eIF3e-mediated translation. Our data uncover critical functions for eIF3d/eIF3e in the hypoxic response and identify a potential means to inhibit stress-induced translation, and potentially plasticity and metastasis, mediated by eIF3e.
Conflict of interest statement
Disclosure and competing interests statement: A patent application has been filed (XW, WO, RZ, HLF) on “Small molecules targeting eIF3e to inhibit tumor growth progression, and metastasis”. Application #:18/839,131. All other authors declare they have no competing interests.
Figures


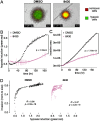
References
-
- Society A.C., Cancer Facts and Figures 2024. 2024, Atlanta: American Cancer Society.
-
- Fabbri L., et al. , The plasticity of mRNA translation during cancer progression and therapy resistance. Nat Rev Cancer, 2021. 21(9): p. 558–577. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
