This is a preprint.
Desiccation-induced DNA damage facilitates drug resistance in Mycobacterium tuberculosis
- PMID: 40501622
- PMCID: PMC12157685
- DOI: 10.1101/2025.06.04.657859
Desiccation-induced DNA damage facilitates drug resistance in Mycobacterium tuberculosis
Abstract
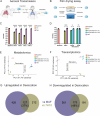
Mycobacterium tuberculosis (Mtb) is an obligate human pathogen that depends on its ability to spread from host-to-host to survive as a species. Yet, knowledge of transmission-specific traits remains lacking. Here, we report the discovery of a specific adaptive response to desiccation, a stress intrinsically linked to the generation of the aerosol droplets within which Mtb transmits. We show that desiccation inflicts oxidative damage and activates Mtb's DNA repair responses but that this repair is imperfect and results in mutations. We further show that activation of these DNA repair responses is accompanied by increased expression of the transcription-coupled repair factor, mfd, but that this expression serves to buffer the fitness cost of specific resistance-conferring mutations in rpoB, the target of the frontline drug rifampin, rather than to facilitate transcription-coupled DNA repair. Silencing mfd during aerosolization impairs survival of strains harboring the rifampin resistance allele S450L. This function is further supported by whole genome sequence data from over 50,000 clinically circulating strains. These studies indicate that Mtb has evolved transmission-specific stress responses that have enabled it to leverage desiccation-induced DNA damage as a potential source of genetic diversification and drug resistance.
Figures

Similar articles
-
Xpert® MTB/RIF assay for extrapulmonary tuberculosis and rifampicin resistance.Cochrane Database Syst Rev. 2018 Aug 27;8(8):CD012768. doi: 10.1002/14651858.CD012768.pub2. Cochrane Database Syst Rev. 2018. Update in: Cochrane Database Syst Rev. 2021 Jan 15;1:CD012768. doi: 10.1002/14651858.CD012768.pub3. PMID: 30148542 Free PMC article. Updated.
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
-
Home treatment for mental health problems: a systematic review.Health Technol Assess. 2001;5(15):1-139. doi: 10.3310/hta5150. Health Technol Assess. 2001. PMID: 11532236
-
Xpert MTB/XDR for detection of pulmonary tuberculosis and resistance to isoniazid, fluoroquinolones, ethionamide, and amikacin.Cochrane Database Syst Rev. 2022 May 18;5(5):CD014841. doi: 10.1002/14651858.CD014841.pub2. Cochrane Database Syst Rev. 2022. PMID: 35583175 Free PMC article.
-
Xpert MTB/RIF Ultra assay for tuberculosis disease and rifampicin resistance in children.Cochrane Database Syst Rev. 2022 Sep 6;9(9):CD013359. doi: 10.1002/14651858.CD013359.pub3. Cochrane Database Syst Rev. 2022. PMID: 36065889 Free PMC article.
References
-
- Goig G. A. et al. , Ecology, global diversity and evolutionary mechanisms in the Mycobacterium tuberculosis complex. Nat Rev Microbiol, (2025). - PubMed
-
- Global tuberculosis report 2024. Geneva: World Health Organization. (2024).
-
- Ernst J. D., The immunological life cycle of tuberculosis. Nat Rev Immunol 12, 581–591 (2012). - PubMed
-
- Cambier C. J., Falkow S., Ramakrishnan L., Host evasion and exploitation schemes of Mycobacterium tuberculosis. Cell 159, 1497–1509 (2014). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
