This is a preprint.
Transcriptional Repression of reaper by Stand Still Safeguards Female Germline Development in Drosophila
- PMID: 40501627
- PMCID: PMC12157695
- DOI: 10.1101/2025.05.17.654630
Transcriptional Repression of reaper by Stand Still Safeguards Female Germline Development in Drosophila
Abstract
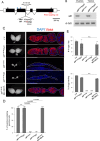
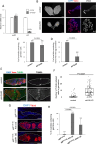
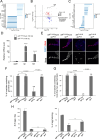
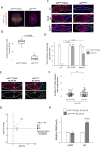
Apoptosis plays a central role in shaping tissues and preserving cellular integrity across developmental stages. In the germline, its precise regulation is critical to ensure both the elimination of aberrant cells and the maintenance of reproductive capacity. However, the molecular mechanisms that control apoptotic susceptibility in germline cells remain poorly defined. Here, we identify stand still (stil) as a female germline-specific regulator of apoptosis in Drosophila. Loss of stil leads to near-complete depletion of germline cells at the time of eclosion, associated with upregulation of the pro-apoptotic gene reaper (rpr) and activation of caspase-dependent cell death. Reporter assays in S2 cells show that Stil directly represses rpr transcription through its N-terminal BED-type zinc finger domain. Despite the absence of stil, undifferentiated germline cells remain resistant to apoptosis. Analysis of publicly available chromatin data reveals that the rpr locus in these cells resides in a closed, H3K9me3-enriched chromatin state, suggesting a Stil-independent mode of transcriptional silencing. Together, our findings uncover two distinct mechanisms that protects the female germline from rpr-dependent apoptosis: Stil-mediated transcriptional repression that operates in both undifferentiated and differentiated germline cells, and an additional chromatin-based silencing mechanism that functions specifically in undifferentiated cells. This work provides new insights into the interplay between transcriptional and chromatin-based regulations that maintain germline cell identity and survival.
Conflict of interest statement
Competing Interest Statement The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Immunogenicity and seroefficacy of pneumococcal conjugate vaccines: a systematic review and network meta-analysis.Health Technol Assess. 2024 Jul;28(34):1-109. doi: 10.3310/YWHA3079. Health Technol Assess. 2024. PMID: 39046101 Free PMC article.
-
Home treatment for mental health problems: a systematic review.Health Technol Assess. 2001;5(15):1-139. doi: 10.3310/hta5150. Health Technol Assess. 2001. PMID: 11532236
-
Evidence That Dmrta2 Acts through Repression of Pax6 in Cortical Patterning and Identification of a Mutation Impairing DNA Recognition Associated with Microcephaly in Human.eNeuro. 2025 Jun 20;12(6):ENEURO.0377-24.2025. doi: 10.1523/ENEURO.0377-24.2025. Print 2025 Jun. eNeuro. 2025. PMID: 40541527 Free PMC article.
-
Reading aids for adults with low vision.Cochrane Database Syst Rev. 2018 Apr 17;4(4):CD003303. doi: 10.1002/14651858.CD003303.pub4. Cochrane Database Syst Rev. 2018. PMID: 29664159 Free PMC article.
-
Interventions targeted at women to encourage the uptake of cervical screening.Cochrane Database Syst Rev. 2021 Sep 6;9(9):CD002834. doi: 10.1002/14651858.CD002834.pub3. Cochrane Database Syst Rev. 2021. PMID: 34694000 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
