This is a preprint.
Nuclear Phase Separation Drives NPM1-mutant Acute Myeloid Leukemia
- PMID: 40501735
- PMCID: PMC12154763
- DOI: 10.1101/2025.05.23.655671
Nuclear Phase Separation Drives NPM1-mutant Acute Myeloid Leukemia
Abstract
During cancer development, mutations promote gene expression changes that cause transformation. Leukemia is frequently associated with aberrant HOXA expression driven by translocations in nucleoporin genes or KMT2A, and mutations in NPM1. How disparate mutations converge on this regulatory pathway is not understood. Here we demonstrate that mutant NPM1 (NPM1c) forms nuclear condensates in multiple human cell lines, mouse models, and primary patient samples. We show NPM1c phase separation is necessary and sufficient to coordinate the recruitment of NUP98 and KMT2A to condensates. Through extensive mutagenesis and pharmacological destabilization of phase separation, we find that NPM1c condensates are necessary for regulating gene expression, promoting in vivo expansion, and maintaining the undifferentiated leukemic state. Finally, we show that nucleoporin and KMT2A fusion proteins form condensates that are biophysically indistinguishable from NPM1c condensates. Together, these data define a new condensate underlying leukemias that we term coordinating bodies (C-bodies), and propose C-bodies as a therapeutic vulnerability.
Figures
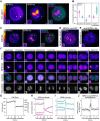
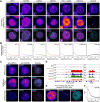
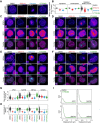
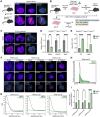
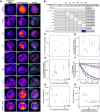
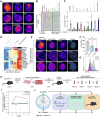
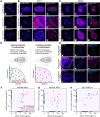
References
-
- Grisendi S., Mecucci C., Falini B., and Pandolfi P.P. (7/2006). Nucleophosmin and cancer. Nat. Rev. Cancer 6, 493–505. - PubMed
-
- Mitrea D.M., Cika J.A., Guy C.S., Ban D., Banerjee P.R., Stanley C.B., Nourse A., Deniz A.A., and Kriwacki R.W. (2016). Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. Elife 5. 10.7554/eLife.13571. - DOI - PMC - PubMed
-
- Yao Z., Duan S., Hou D., Wang W., Wang G., Liu Y., Wen L., and Wu M. (2010). B23 acts as a nucleolar stress sensor and promotes cell survival through its dynamic interaction with hnRNPU and hnRNPA1. Oncogene 29, 1821–1834. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
