This is a preprint.
Transcription factors SP5 and SP8 drive primary cilia formation
- PMID: 40501818
- PMCID: PMC12157413
- DOI: 10.1101/2025.06.03.657415
Transcription factors SP5 and SP8 drive primary cilia formation
Update in
-
Transcription factors SP5 and SP8 drive primary cilia formation in mammalian embryos.Science. 2025 Aug 28;389(6763):eadt5663. doi: 10.1126/science.adt5663. Epub 2025 Aug 28. Science. 2025. PMID: 40875857
Abstract
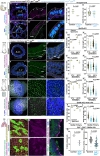
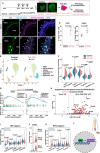
While specific transcription factors are known to regulate cell fate decisions, the degree to which they can stimulate formation of specific cell organelles is less clear. We used a multi-omics comparison of the transcriptomes of ciliated and non-ciliated embryonic cells to identify transcription factors upregulated in ciliated cells, and conditional genetics in mouse embryos and stem cells to demonstrate that SP5/8 regulate cilia formation and gene expression. In Sp5/8 mutant embryos primary and motile cilia are shorter than normal and reduced in number across cell types, contributing to situs inversus and hydrocephalus. Moreover, expression of SP8 is sufficient to induce primary cilia in unciliated cells. This work opens new avenues for studying cilia assembly using stem cell models and offers new insights into human ciliopathies.
Conflict of interest statement
Competing interests: The authors declare no competing financial interests.
Figures



References
-
- H. M. Mitchison, E. M. Valente, Motile and non-motile cilia in human pathology: from function to phenotypes. J Pathol 241, 294–309 (2017). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
