This is a preprint.
Loss of tumor suppressor p53 upregulates stem cell factor SOX9 via Notch signaling
- PMID: 40501825
- PMCID: PMC12154799
- DOI: 10.1101/2025.05.27.656408
Loss of tumor suppressor p53 upregulates stem cell factor SOX9 via Notch signaling
Abstract
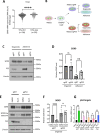
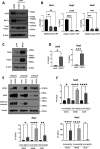
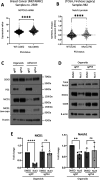
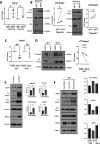
Basal-like breast cancer (BLBC) consists of the majority of triple-negative breast cancer subtype that has a higher degree of cellular plasticity owing to a greater number of stem-like cancer cells compared to other subtypes1. BLBCs are thought to originate from the luminal progenitor cells despite their prominent basal-cell features. SOX9 is a key transcription factor which is expressed selectively in estrogen receptor-negative luminal progenitors in postnatal mammary glands. During BLBC progression, SOX9 upregulation is required for the de-differentiation of luminal cells to multipotent fetal mammary stem cell-like states, which are crucial for the malignant progression of BLBC. However, the mechanism driving SOX9 upregulation in BLBC remains unclear. Since p53 is mutated in nearly 90% of the BLBC and is considered as an early event in the BLBC transformation, we hypothesized that p53 loss could contribute to SOX9 upregulation during BLBC progression. Using primary mammary cell organoids, we showed that p53 loss not only induced the SOX9 expression but also stabilized the half-life of SOX9 protein. We further identified that p53 loss increased Psen2 expression to activate the Notch1 signaling, which induced SOX9 expression. Together, our work identified a molecular mechanism allowing loss of tumor suppressor p53 to coopt the cell fate determinant SOX9 to drive de-differentiation in BLBC.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures
References
-
- Van Keymeulen A. et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature 479, 189–193 (2011). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
