This is a preprint.
Proximity biotinylation at the host- Shigella interface reveals UFMylation as an antibacterial pathway
- PMID: 40501833
- PMCID: PMC12154702
- DOI: 10.1101/2025.05.29.656827
Proximity biotinylation at the host- Shigella interface reveals UFMylation as an antibacterial pathway
Abstract
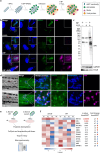
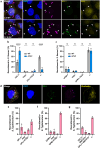
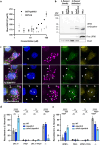
Host cells contest invasion by intracellular bacterial pathogens with multiple strategies that recognise and / or damage the bacterial surface. To identify novel host defence factors targeted to intracellular bacteria, we developed a versatile proximity biotinylation approach coupled to quantitative mass spectrometry that maps the host-bacterial interface during infection. Using this method, we discovered that intracellular Shigella and Salmonella become targeted by UFM1-protein ligase 1 (UFL1), an E3 ligase that catalyses the covalent attachment of Ubiquitin-fold modifier 1 (UFM1) to target substrates in a process called UFMylation. We show that Shigella antagonises UFMylation in a dual manner: first, using its lipopolysaccharide (LPS) to shield from UFL1 recruitment; second, preventing UFM1 decoration by the bacterial effector IpaH9.8. Absence of UFMylation leads to an increase of bacterial burden in both human cells and zebrafish larvae, suggesting that UFMylation is a highly conserved antibacterial pathway. Contrary to canonical ubiquitylation, the protective role of UFMylation is independent of autophagy. Altogether, our proximity mapping of the host-bacterial interface identifies UFMylation as an ancient antibacterial pathway and holds great promise to reveal other cell-autonomous immunity mechanisms.
Keywords: E3 ligases; Shigella; UFMylation; bacterial infection; cell-autonomous immunity; proximity biotinylation; ubiquitin-like systems.
Figures

References
-
- Ray K. et al. Tracking the dynamic interplay between bacterial and host factors during pathogen-induced vacuole rupture in real time. Cell Microbiol 12, 545–556 (2010). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
