This is a preprint.
TITAN-RNA: A hybrid-capture sequencing panel detects known and unknown Flaviviridae for diagnostics and vector surveillance
- PMID: 40501929
- PMCID: PMC12157608
- DOI: 10.1101/2025.06.08.658352
TITAN-RNA: A hybrid-capture sequencing panel detects known and unknown Flaviviridae for diagnostics and vector surveillance
Abstract
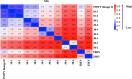
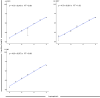
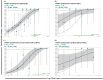
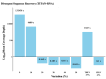
Clinical testing and public health surveillance can be significantly improved by incorporating sequencing-based molecular detection and subtyping for real-time monitoring of virus evolution. With phylogenetic analysis used for speciation and variant subtyping, target analyte specificity can be relaxed well beyond typical parameters acceptable in PCR-based diagnostics. Hybrid capture is a promising way to enrich large numbers of sequences with maximal flexibility, using standard molecular biology laboratory equipment and small benchtop sequencers. Here, we report the development and bench validation of a hybrid capture based next-generation sequencing diagnostic panel for all known viral tick-borne pathogens, TITAN-RNA. Based on systematic testing with simulated novel viruses and field samples, we determined a 10% tolerance for evenly distributed mutations or 27% tolerance for naturally occurring viral divergence. The TITAN-RNA extrapolated limit of detection in blood is 19.1 genome copies by complementary log-log analysis, and linearity performance (R2 ≥ 0.99) is amenable for its use as a quantitative assay. As proof of principle for public health surveillance and evolutionary studies, we report two putatively novel segmented Flavi-like viruses in New York State, USA, identified from the invasive Haemaphysalis longicornis tick.
Conflict of interest statement
Conflict of Interest The authors declare no conflicts.
Figures


Similar articles
-
Rapid, point-of-care antigen tests for diagnosis of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2022 Jul 22;7(7):CD013705. doi: 10.1002/14651858.CD013705.pub3. Cochrane Database Syst Rev. 2022. PMID: 35866452 Free PMC article.
-
Diagnostic test accuracy and cost-effectiveness of tests for codeletion of chromosomal arms 1p and 19q in people with glioma.Cochrane Database Syst Rev. 2022 Mar 2;3(3):CD013387. doi: 10.1002/14651858.CD013387.pub2. Cochrane Database Syst Rev. 2022. PMID: 35233774 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Antibody tests for identification of current and past infection with SARS-CoV-2.Cochrane Database Syst Rev. 2022 Nov 17;11(11):CD013652. doi: 10.1002/14651858.CD013652.pub2. Cochrane Database Syst Rev. 2022. PMID: 36394900 Free PMC article.
-
TICKHUNTER: A Targeted Hybridization-Capture Sequencing Approach for the Detection and Characterization of Tick-borne Pathogens and Blood Meals.bioRxiv [Preprint]. 2025 Jun 6:2025.06.06.658355. doi: 10.1101/2025.06.06.658355. bioRxiv. 2025. PMID: 40501899 Free PMC article. Preprint.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
