This is a preprint.
CTCF maintains pericentromere function and mitotic fidelity
- PMID: 40502007
- PMCID: PMC12157408
- DOI: 10.1101/2025.05.30.657091
CTCF maintains pericentromere function and mitotic fidelity
Update in
-
CTCF maintains centromere function and mitotic fidelity.J Cell Sci. 2026 Jan 5:jcs.264181. doi: 10.1242/jcs.264181. Online ahead of print. J Cell Sci. 2026. PMID: 41486985
Abstract
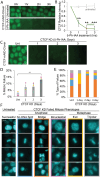
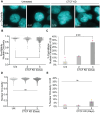
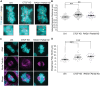
In mitosis the duplicated genome is aligned and accurately segregated between daughter nuclei. CTCF is a chromatin looping protein in interphase with an unknown role in mitosis. We previously published data showing that CTCF constitutive knockdown causes mitotic failure, but the mechanism remains unknown. To determine the role of CTCF in mitosis, we used a CRISPR CTCF auxin inducible degron cell line for rapid degradation. CTCF degradation for 3 days resulted in increased failure of mitosis and decreased circularity in post-mitotic nuclei. Upon CTCF degradation CENP-E is still recruited to the kinetochore and there is a low incidence of polar chromosomes which occur upon CENP-E inhibition. Instead, immunofluorescence imaging of mitotic spindles reveals that CTCF degradation causes increased intercentromere distances and a wider and more disorganized metaphase plate, a disruption of key functions of the pericentromere. These results are similar to partial loss of cohesin, an established component of the pericentromere. Thus, we reveal that CTCF is a key maintenance factor of pericentromere function, successful mitosis, and post-mitotic nuclear shape.
Conflict of interest statement
Competing interests The authors declare not competing interests.
Figures

References
-
- Barisic M, Aguiar P, Geley S, and Maiato H (2014). Kinetochore motors drive congression of peripheral polar chromosomes by overcoming random arm-ejection forces. Nat Cell Biol 16, 1249–1256. - PubMed
Publication types
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
