This is a preprint.
Klf4-dependent pivotal role of smooth muscle-derived Gli1 + lineage progenitor cells in lung cancer progression
- PMID: 40502062
- PMCID: PMC12157427
- DOI: 10.1101/2025.05.30.657094
Klf4-dependent pivotal role of smooth muscle-derived Gli1 + lineage progenitor cells in lung cancer progression
Abstract
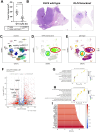
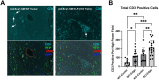
Lung cancer remains the leading cause of cancer-related deaths worldwide. Despite the remarkable efficacy of immune checkpoint inhibitors, only a subset of lung adenocarcinoma (LUAD) patients respond and many eventually progress, underscoring the need to develop novel combination therapies. The interaction between cancer cells and the tumor microenvironment (TME), a complex niche including cells of the innate and adaptive immune system, cancer-associated fibroblasts (CAFs), vascular cells, and extracellular matrix (ECM) plays a vital role in tumorigenesis and cancer progression. Using fate mapping, we previously identified a novel subpopulation of resident vascular stem cells derived from vascular smooth muscle cells, designated AdvSca1-SM (vascular Adventitial location, Stem cell antigen-1 expression, SMooth muscle origin) cells. AdvSca1-SM cells are the predominant cell type responding to vessel wall dysfunction and their selective differentiation to myofibroblasts promotes vascular fibrosis. SMC-to-AdvSca1-SM cell reprogramming is dependent on SMC induction of the transcription factor KLF4, and KLF4 is essential for maintenance of the stem cell phenotype. The function of AdvSca1-SM cells in LUAD tumorigenesis has not been explored. Using an orthotopic immunocompetent mouse model of LUAD, we demonstrate that AdvSca1-SM cells are a major component of lung tumors, significantly contributing to cancer associated fibroblasts (CAFs). Compared to AdvSca1-SM cells, CAFs have altered ECM gene expression with a reduction of a stemness signature. AdvSca1-SM-specific genetic ablation of Klf4 altered their phenotype resulting in inhibition of communication between cancer cells and CAFs, decreases in innate immunosuppressive populations, and increased T cell infiltration into the tumor. Targeting this population may represent a novel strategy to improve the response to immunotherapy.
Figures




References
-
- Sezer A., Kilickap S., Gümüş M., Bondarenko I., Özgüroğlu M., Gogishvili M., Turk H.M., Cicin I., Bentsion D., Gladkov O., et al. (2021). Cemiplimab monotherapy for first-line treatment of advanced non-small-cell lung cancer with PD-L1 of at least 50%: a multicentre, open-label, global, phase 3, randomised, controlled trial. Lancet 397, 592–604. 10.1016/S0140-6736(21)00228-2. - DOI - PubMed
-
- Gogishvili M., Melkadze T., Makharadze T., Giorgadze D., Dvorkin M., Penkov K., Laktionov K., Nemsadze G., Nechaeva M., Rozhkova I., et al. (2022). Cemiplimab plus chemotherapy versus chemotherapy alone in non-small cell lung cancer: a randomized, controlled, double-blind phase 3 trial. Nat Med 28, 2374–2380. 10.1038/s41591-022-01977-y. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
