This is a preprint.
Movi Color: fast and accurate long-read classification with the move structure
- PMID: 40502105
- PMCID: PMC12154825
- DOI: 10.1101/2025.05.22.655637
Movi Color: fast and accurate long-read classification with the move structure
Abstract
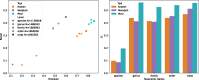
The number of reference genomes is rapidly increasing, thanks to advances in long-read sequencing and assembly. While these collections can improve the sensitivity and specificity of classification methods, this requires highly efficient compressed indexes. K-mer-based approaches like Kraken 2 are efficient but limit the analysis to a fixed k-mer length. This is hard for the user to set ahead of time, and suboptimal settings can harm sensitivity and specificity. Methods that use compressed full-text indexes like SPUMONI2 and Cliffy lift this constraint, but are less efficient than k-mer-based tools. Further, these methods either cannot report a full listing of genomes where a match occurs, or cannot scale to large reference databases. We propose new methods and algorithms that use compressed full-text indexes to enable multi-class and taxonomic classification. Unlike past compressed-indexing methods for classification, ours uses the move structure, which is extremely fast thanks to its locality of reference. Our method, called Movi Color, augments the main table of the Movi index. Specifically, Movi Color assigns a "color" to each run of the Burrows-Wheeler Transform according to the subset of genomes from which the run suffixes originated. When the reference is highly repetitive - as is typical when indexing pangenomes or reference databases - only certain colors occur, creating opportunities to compress the index. For species-level classification, Movi Color achieves over 1.6× higher precision and 2× higher recall than Kraken 2 and Metabuli. At the genus level, it achieves 70% higher precision and 80% higher recall. Movi Color's read processing time is 7-20× faster than Metabuli and is a comparable to Kraken 2. Although Movi Color uses more memory than both Kraken 2 and Metabuli, its speed-accuracy trade-off makes it well-suited for real-time or high-throughput scenarios.
Keywords: Applied computing; Comparative genomics; Compressed indexing; Computational genomics; Pangenomics.
Figures


Similar articles
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Diagnostic test accuracy and cost-effectiveness of tests for codeletion of chromosomal arms 1p and 19q in people with glioma.Cochrane Database Syst Rev. 2022 Mar 2;3(3):CD013387. doi: 10.1002/14651858.CD013387.pub2. Cochrane Database Syst Rev. 2022. PMID: 35233774 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Antidepressants for pain management in adults with chronic pain: a network meta-analysis.Health Technol Assess. 2024 Oct;28(62):1-155. doi: 10.3310/MKRT2948. Health Technol Assess. 2024. PMID: 39367772 Free PMC article.
-
Methods for blood loss estimation after vaginal birth.Cochrane Database Syst Rev. 2018 Sep 13;9(9):CD010980. doi: 10.1002/14651858.CD010980.pub2. Cochrane Database Syst Rev. 2018. PMID: 30211952 Free PMC article.
References
-
- Ahmed Omar, Boucher Christina, and Langmead Ben. Cliffy: robust 16s rrna classification based on a compressed lca index. bioRxiv, pages 2024–05, 2024.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
