This is a preprint.
OrganoidChip+: a Microfluidic Platform for Culturing, Staining, Immobilization, and High-Content Imaging of Adult Stem Cell-Derived Organoids
- PMID: 40502189
- PMCID: PMC12157677
- DOI: 10.1101/2025.06.02.657499
OrganoidChip+: a Microfluidic Platform for Culturing, Staining, Immobilization, and High-Content Imaging of Adult Stem Cell-Derived Organoids
Update in
-
A microfluidic platform for culturing and high-content imaging of adult stem cell-derived organoids.Sci Rep. 2025 Nov 17;15(1):40162. doi: 10.1038/s41598-025-23883-2. Sci Rep. 2025. PMID: 41249220 Free PMC article.
Abstract
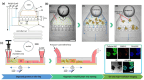
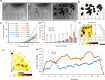
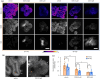
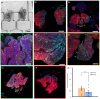
High-content imaging (HCI) and analysis are the keys for advancing our understanding of the science behind organogenesis. To this end, culturing adult stem cell-derived organoids (ASOs) in a platform that also enables live imaging, staining, immobilization, and fast high-resolution imaging is crucial. However, existing platforms only partially satisfy these requirements. In this study, we present the OrganoidChip+, an all-in-one microfluidic device designed to integrate both culturing and HCI of ASOs all within one platform. We previously developed the OrganoidChip as a robust imaging tool. Now, the OrganoidChip+ incorporates several additional features for culturing organoids in addition to fluorescence staining and imaging without the need for sample transfer. The organoids grown within a culture chamber are stained and then transferred to immobilization chambers for blur-free, high-resolution imaging at predetermined locations. We cultured adult stem cell-derived intestinal organoids in the chip for 7 days and tracked growth rates of each organoid using intermittent brightfield images, followed by multiple image-based assays, including viability assay using widefield fluorescence imaging, a redox ratio assay using label-free, two-color, two-photon microscopy, and immunofluorescence assays using confocal microscopy. These assays serve as proof-of-concept to showcase the chip's capabilities in HCI of ASOs. Organoids cultured in the chip exhibited superior average growth rates over those in traditional Matrigel dome cultures, off-chip. Viability and redox ratio measurements of on-chip organoids were comparable or slightly better than their off-chip counterparts. Confocal imaging further confirmed that the OrganoidChip+ supports robust organoid culture while enabling detailed, high-resolution analysis. This all-in-one platform holds great potential for advancing ASO-based research, offering a scalable and cost-effective solution for HCI and analysis in organogenesis, drug screening, and disease modeling.
Conflict of interest statement
Conflict of interest A.B., K.M., S.M., E.H., and A.H. are listed as inventors on a pending U.S. patent application (PCT/US24/13330) directly related to the platform described in this work, which has been licensed to vivoVerse, LLC. Additionally, A.B., S.M., and E.H. hold equity in vivoVerse, LLC. These disclosures have been reviewed and managed according to institutional policies addressing potential conflicts of interest in research. K.A. is a co-founder of LifEngine Animal Health and 3D Health Solutions and serves as a consultant for Ceva Animal Health, Bioiberica, LifeDiagnostics, Antech Diagnostics, Deerland Probiotics, Christian Hansen Probiotics, Purina, and Mars. J.P.M. is a co-founder of LifEngine Animal Health (LEAH) and 3D Health Solutions and serves as a consultant for Ceva Animal Health, Ethos Animal Health, LifEngine Animal Health and Boehringer Ingelheim. C.Z. is the Director of Research and Product Development at 3D Health Solutions.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
