This is a preprint.
A Biological-Systems-Based Analyses Using Proteomic and Metabolic Network Inference Reveals Mechanistic Insights into Hepatic Lipid Accumulation: An IMI-DIRECT Study
- PMID: 40502600
- PMCID: PMC12155020
- DOI: 10.1101/2025.06.02.25328773
A Biological-Systems-Based Analyses Using Proteomic and Metabolic Network Inference Reveals Mechanistic Insights into Hepatic Lipid Accumulation: An IMI-DIRECT Study
Abstract
Objective: To delineate organ-specific and systemic drivers of metabolic dysfunction-associated steatotic liver disease (MASLD), we applied integrative causal inference across clinical, imaging, and proteomic domains in individuals with and without type 2 diabetes (T2D).
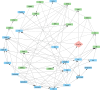
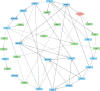
Research design and methods: We used Bayesian network analyses to quantify causal pathways linking adipose distribution, glycemia, and insulin dynamics with fatty liver using data from the IMI-DIRECT prospective cohort study. Measurements were made of glucose and insulin dynamics (using frequently-sampled metabolic challenge tests), MRI-derived abdominal and liver fat content, serological biomarkers, and Olink plasma proteomics from 331 adults with new-onset T2D and 964 adults free from diabetes at enrolment. The common protocols used in these two cohorts provided the opportunity for replication analyses to be performed. When the direction of the effect could not be determined with high probability through Bayesian networks, complementary two-sample Mendelian randomization (MR) was employed.
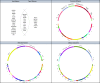
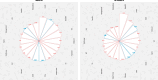
Results: High basal insulin secretion rate (BasalISR) was identified as the primary causal driver of liver fat accumulation in both diabetes and non-diabetes. Excess visceral adipose tissue (VAT) was bidirectionally associated with liver fat, indicating a self-reinforcing metabolic loop. Basal insulin clearance (Clinsb) worsened as a consequence of liver fat accumulation to a greater degree before the onset of T2D. Out of 446 analysed proteins, 34 mapped to these metabolic networks and 27 were identified in the non-diabetes network, 18 in the diabetes network, and 11 were common between the two networks. Key proteins directly associated with liver fat included GUSB, ALDH1A1, LPL, IGFBP1/2, CTSD, HMOX1, FGF21, AGRP, and ACE2. Sex-stratified analyses revealed distinct proteomic drivers: GUSB and LEP were most predictive of liver fat in females and males, respectively.
Conclusions: Basal insulin hypersecretion is a modifiable, causal driver of MASLD, particularly prior to glycaemic decompensation. Our findings highlight a multifactorial, sex- and disease-stage-specific proteo-metabolic architecture of hepatic steatosis. Proteins such as GUSB, ALDH1A1, LPL, and IGFBPs warrant further investigation as potential biomarkers or therapeutic targets for MASLD prevention and treatment.
Conflict of interest statement
DECLARATION OF INTERESTS AD works for Novo Nordisk Research Centre Oxford. SB has ownerships in Intomics A/S, Hoba Therapeutics Aps, Novo Nordisk A/S, Lundbeck A/S, and managing board memberships in Proscion A/S and Intomics A/S. MR owns stock in Novo Nordisk A/S. MMcC is an employee of Genentech and a holder of Roche stock. Within the past five years, PWF has received consulting honoraria from Eli Lilly Inc., Novo Nordisk Foundation, Novo Nordisk A/S, UBS, Qatar Foundation, and Zoe Ltd. PWF was also an employee of the Novo Nordisk Foundation (2021–2024). PWF has also received investigator-initiated grants (paid to institution) from numerous pharmaceutical companies as part of the Innovative Medicines Initiative of the European Union.
Figures
References
-
- En Li Cho E, Ang CZ, Quek J, Fu CE, Lim LKE, Heng ZEQ, et al. Global prevalence of non-alcoholic fatty liver disease in type 2 diabetes mellitus: an updated systematic review and meta-analysis. Gut. 2023;72(11):2138–48. - PubMed
-
- Hyogo H, Yamagishi S, Maeda S, Kimura Y, Ishitobi T, Chayama K. Increased insulinogenic index is an independent determinant of nonalcoholic fatty liver disease activity score in patients with normal glucose tolerance. Dig Liver Dis. 2012;44(11):935–9. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
