Exploring the role of neutrophils in inflammatory pain hypersensitivity via single-cell transcriptome profiling
- PMID: 40503232
- PMCID: PMC12151834
- DOI: 10.3389/fimmu.2025.1552993
Exploring the role of neutrophils in inflammatory pain hypersensitivity via single-cell transcriptome profiling
Abstract
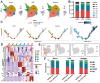
Introduction: Myeloid CD11b+ cells are crucial mediators in post-operative and CFA-induced inflammation, but their role in pain, particularly the role of neutrophils, is still debated. This study employs single-cell RNA sequencing (scRNA-seq) to analyze CD11b+ cell composition in mice after surgery and CFA treatment and investigates the effects and mechanisms of Nicotinamide N-oxide (NAMO) on neutrophils and pain.
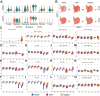
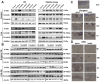
Methods: scRNA-seq was used to analyze the transcriptomes of CD11b+ cells in murine models of post-operative and CFA-induced inflammation. Using comprehensive bioinformatics techniques, we identified distinct cell subpopulations and characterized their gene expression profiles and functional attributes. Based on these analyses, NAMO was selected to intervene in neutrophil differentiation and maturation. The role of the CXCR2 target gene and NAMO in modulating post-operative and inflammatory pain was then evaluated, exploring potential mechanisms.
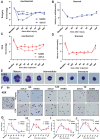
Results: scRNA-seq revealed a significant increase in neutrophils and a decrease in monocytes among CD11b+ cells following surgery and CFA treatment. Neutrophils comprised seven subpopulations at various differentiation stages from immature to mature. Given the high expression of CXCR2 in neutrophils, we used the CXCR2 inhibitor NAMO to suppress neutrophil differentiation and maturation, which subsequently alleviated post-operative and CFA-induced pain in mice. Proteomics analysis showed that NAMO treatment significantly reduced the expression of S100b and CaMKIIβ proteins in mouse neutrophils.
Discussion: Following surgery and CFA treatment, mature neutrophils were significantly elevated. The CXCR2 antagonist NAMO alleviated post-surgical and CFA-induced pain by inhibiting neutrophil differentiation and maturation. These findings offer novel approaches for pain prevention and treatment.
Keywords: CXCR2; NAMO; inflammation; neutrophil; pain.
Copyright © 2025 Ding, Liu, Zeng, Liu, Zhang, Li, Zhou, Su and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

