PD-1 is conserved from sharks to humans: new insights into PD-1, PD-L1, PD-L2, and SHP-2 evolution
- PMID: 40503235
- PMCID: PMC12151841
- DOI: 10.3389/fimmu.2025.1573492
PD-1 is conserved from sharks to humans: new insights into PD-1, PD-L1, PD-L2, and SHP-2 evolution
Abstract
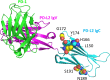
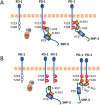
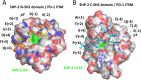
Programmed cell death protein 1 (PD-1) is an immune checkpoint molecule until recently believed to exist only in tetrapod species. However, together with a very recent study dedicated to the CD28/CTLA4 molecule family, this study-using database information-identifies the PD-1 gene in both bony and cartilaginous fish, while being the first to present a detailed molecular analysis of the evolution of PD-1 and its ligands. Conserved sequence motifs imply an ancient origin of PD-1's binding modes to its extracellular ligand PD-L1 and its intracellular ligand Src homology region 2 domain-containing phosphatase-2 (SHP-2), and also of its N116 glycosylation motif-a less well known PD-1 feature-important for binding galectins. The PD-1 cytoplasmic tail binds SHP-2 by two motifs, defined as an immunoreceptor tyrosine-based inhibitory motif (ITIM) and immunoreceptor tyrosine-based switch motif (ITSM), but sequence conservation patterns show that these definitions warrant a discussion. As in mammals, PD-1 transcripts in fish could be found co-expressed with markers of regulatory and exhausted T cells, suggesting a similar immune checkpoint function. Agreeing with previous reports, the PD-L1/PD-L2 gene duplication was only found in tetrapod species, while we newly discovered that features that consistently distinguish the two molecules are PD-L2 IgC domain motifs. Among PD-L1 (the name given to the single PD-L ancestral molecule) of many ray-finned fish, conservation of a very long cytoplasmic tail motif supports previous claims that PD-L1 cytoplasmic tails may have a function. Surprisingly, we found a gene similar to SHP-2-that we named SHP-2-like (SHP-2L)-to be conserved from sharks to mammals, although lost or inactivated in higher primates and rodents. SHP-2L is expected to bind PD-1 similar to SHP-2. This comparative analysis of PD-1 and its interacting molecules across jawed vertebrates highlights conserved immune checkpoint features while revealing new insights and lineage-specific adaptations.
Keywords: PD-1; PD-L1; PD-L2; SHP-1; SHP-2; SHP-2L; evolution; fish.
Copyright © 2025 Kondo, Kondo, Nabeshima, Nishikimi, Ishida, Shigeoka and Dijkstra.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision
Figures






Similar articles
-
Interaction of SHP-2 SH2 domains with PD-1 ITSM induces PD-1 dimerization and SHP-2 activation.Commun Biol. 2020 Mar 17;3(1):128. doi: 10.1038/s42003-020-0845-0. Commun Biol. 2020. PMID: 32184441 Free PMC article.
-
SHP-1 and SHP-2 associate with immunoreceptor tyrosine-based switch motif of programmed death 1 upon primary human T cell stimulation, but only receptor ligation prevents T cell activation.J Immunol. 2004 Jul 15;173(2):945-54. doi: 10.4049/jimmunol.173.2.945. J Immunol. 2004. PMID: 15240681
-
The structural features that distinguish PD-L2 from PD-L1 emerged in placental mammals.J Biol Chem. 2020 Apr 3;295(14):4372-4380. doi: 10.1074/jbc.AC119.011747. Epub 2019 Dec 27. J Biol Chem. 2020. PMID: 31882544 Free PMC article.
-
Structural Biology of the Immune Checkpoint Receptor PD-1 and Its Ligands PD-L1/PD-L2.Structure. 2017 Aug 1;25(8):1163-1174. doi: 10.1016/j.str.2017.06.011. Structure. 2017. PMID: 28768162 Review.
-
Targeting the programmed death-1 pathway in lymphoid neoplasms.Cancer Treat Rev. 2017 Mar;54:99-109. doi: 10.1016/j.ctrv.2017.01.009. Epub 2017 Feb 11. Cancer Treat Rev. 2017. PMID: 28242522 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

