SARS-CoV-2 nsp15 enhances viral virulence by subverting host antiviral defenses
- PMID: 40504150
- PMCID: PMC12184426
- DOI: 10.1073/pnas.2426528122
SARS-CoV-2 nsp15 enhances viral virulence by subverting host antiviral defenses
Abstract
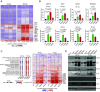
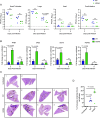
SARS-CoV-2 encodes numerous virulence factors, yet their precise mechanisms of action remain unknown. We provide evidence that the SARS-CoV-2 nonstructural protein 15 (nsp15) enhances viral virulence by suppressing the production of viral double-stranded (dsRNA), a potent inducer of antiviral signaling. The viral variants lacking nsp15 endoribonuclease activity elicited higher innate immune responses and exhibited reduced replication in human stem cell-derived lung alveolar type II epithelial cells, as well as in the lungs of infected hamsters. Consistently, these variants caused significantly less weight loss and mortality compared to wild-type (WT) virus in K18-hACE2 mice. Mechanistically, the cells infected with nsp15 mutants accumulated more viral dsRNA, causing enhanced stimulation of the interferon pathway. Chemical inhibition of interferon signaling dampened immune responses to nsp15 mutants and restored their replication to levels similar to the WT virus. These findings indicate that the endoribonuclease activity of nsp15 contributes to viral virulence by limiting the accumulation of viral dsRNA, thereby allowing robust replication with reduced activation of the host innate immune response.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures



References
MeSH terms
Substances
Grants and funding
- HL163494/HHS | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- S10 OD026983/OD/NIH HHS/United States
- R01 AI159945/AI/NIAID NIH HHS/United States
- T32 AI007508/AI/NIAID NIH HHS/United States
- T32 HL007035/HL/NHLBI NIH HHS/United States
- AI159945/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- HL007035/HHS | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- 75N92020C00005/HL/NHLBI NIH HHS/United States
- AI007508/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- K08 HL163494/HL/NHLBI NIH HHS/United States
- S10 OD030269/OD/NIH HHS/United States
- P01 HL170952/HL/NHLBI NIH HHS/United States
- HL170952/HHS | NIH | National Heart, Lung, and Blood Institute (NHLBI)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

