Pathogen-specific host response in critically ill patients with blood stream infections: a nested case-control study
- PMID: 40505417
- PMCID: PMC12182767
- DOI: 10.1016/j.ebiom.2025.105799
Pathogen-specific host response in critically ill patients with blood stream infections: a nested case-control study
Abstract
Background: Knowledge of the contribution of the pathogen to the heterogeneity of the host response to infection is limited. We aimed to compare the host response in critically ill patients with a bloodstream infection (BSI).
Methods: RNA profiles were determined in blood obtained between one day before and after a positive blood culture. Differential expression and pathway analyses were performed on independent patients' samples by RNA sequencing (discovery) or microarray (validation). Additional patients were included for the discovery and validation of transcriptome classifiers of pathogen-specific BSIs. Twenty biomarkers reflecting key host response pathways were measured in blood.
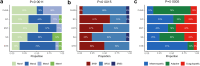
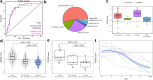
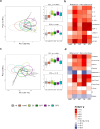
Findings: We included 341 patients, among which 255 with BSI, 25 with viral infection and 61 non-infectious controls. The cultured pathogen explained 41·8% of the blood transcriptomic variance in patients with BSI. Gene set enrichment analysis showed a global resemblance between monomicrobial BSIs caused by Streptococcus, Staphylococcus aureus and Escherichia coli, which were clearly different from BSI caused by coagulase-negative staphylococci or Enterococcus. BSI by Streptococcus was associated with the highest number of differentially expressed genes, indicating strong innate and adaptive immune activation. An eight-gene streptococcal classifier performed well across different Streptococcus species, and was validated in external cohorts. Plasma biomarker profiling showed that E. coli BSI was associated with the strongest response in the cytokine and systemic inflammation domain, and S. aureus BSI with the strongest endothelial cell activation.
Interpretation: The causative pathogen explains a substantial part of the heterogeneity of the host response in critically ill patients with BSI.
Funding: Center for Translational Molecular Medicine and the European Commission.
Keywords: Bacteraemia; Biomarkers; Host response; Intensive care; Transcriptome.
Copyright © 2025 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests Dr. H. Peters-Sengers was supported by the Dutch Kidney Foundation (Nierstichting) postdoc KOLFF grant 19OK009. Dr. van Engelen was supported by an Amsterdam UMC PhD scholarship grant. Dr. van Vught was supported by the Netherlands Organisation for Health Research and Development ZonMW VENI grant 09150161910033. Dr. Schultz is a former research coordinator at Hamilton Medical AG; this role was unrelated to the research presented. Dr Francois received consulting fees from Enlivex and Inotrem, and support for travel from Eagle. Dr. Lombardo is a Takeda employee and owns stock in the company; he received payments from ESAME and Francisco de Votoria University for presentations at Master of Advanced Therapies, and from iBET for accommodation and travel support to attend a thesis defence. Dr Sweeney is an employee of, and stockholder in Inflammatix, Inc. Dr. Bonten is CEO of the European Clinical Research Alliance on Infectious Diseases (Ecraid); in this role he received grants from Sequiris, TechnoPhage, Attea Pharma, Phaxiam and Janssen Vaccines (all paid to Eucraid). Marc Bonten also received grants from Merck, GSK, European Commission, and consulting fees from Merck, GSK and Janssen Vaccines (all paid to UMC Utrecht). Dr. Wiersinga reports grants from the Netherlands Organisation for Health Research and Development (ZonMw), EU/Eurostars and Moderna outside the submitted work. Drs. Simpson and Bolero disclose that they were former employees and shareholders of Immunexpress Inc. Dr. Yager declares that he owns shares of stock and has been granted stock options in Immunexpress Inc; he is a current employee of Immunexpress, Inc. Dr. Cremer received grants from the Centre of Translational Medicine, Health Holland, ZonMW, EU Digital Europe, Presymptom Health Ltd (all paid to UMC Utrecht), he received payment for his participation in the Committee for European Education in Anaesthesiology (paid to UMC Utrecht). Dr. Cremer is Chair of the Science & Innovation Committee of the Dutch Society of Intensive Care, member of the External Project Advisory Committee of the BEATsep Consortium and member of the Scientific Advisory Board of the International Clinical research centre Brno (all without payment). Dr. van der Poll reports grants from Immunexpress, EU/Horizon 2020 (FAIR, Immunosep), the Ministry of Economic Affairs & Health Holland, and the Dutch Thrombosis Foundation, as well as a consultancy with Matisse (all paid to the institution); he is a member of Data Safety Monitoring Board of REMAP-CAP (no payment). Drs. Butler, Reijnders, Uhel, Laterre, Sanchez and Scicluna have no competing interests to declare.
Figures


References
-
- Liu V., Escobar G.J., Greene J.D., et al. Hospital deaths in patients with sepsis from 2 independent cohorts. JAMA. 2014;312(1):90–92. - PubMed
-
- Cajander S., Kox M., Scicluna B.P., et al. Profiling the dysregulated immune response in sepsis: overcoming challenges to achieve the goal of precision medicine. Lancet Respir Med. 2024;12(4):305–322. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

