BRAF/MEK inhibition induces cell state transitions boosting immune checkpoint sensitivity in BRAFV600E-mutant glioma
- PMID: 40505659
- PMCID: PMC12208339
- DOI: 10.1016/j.xcrm.2025.102183
BRAF/MEK inhibition induces cell state transitions boosting immune checkpoint sensitivity in BRAFV600E-mutant glioma
Abstract
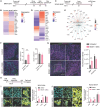
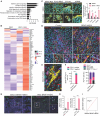
Resistance to v-raf murine sarcoma viral oncogene homolog B1 (BRAF) plus mitogen-activated protein kinase kinase (MEK) inhibition (BRAFi+MEKi) in BRAFV600E-mutant gliomas drives rebound, progression, and high mortality, yet it remains poorly understood. This study addresses the urgent need to develop treatments for BRAFi+MEKi-resistant glioma using preclinical mouse models and patient-derived materials. BRAFi+MEKi reveals glioma plasticity by heightening cell state transitions along glial differentiation trajectories, giving rise to astrocyte- and immunomodulatory oligodendrocyte (OL)-like states. PD-L1 upregulation in OL-like cells links cell state transitions to immune evasion, possibly orchestrated by Galectin-3. BRAFi+MEKi induces interferon response signatures, tumor infiltration, and suppression of T cells. Combining BRAFi+MEKi with immune checkpoint inhibition enhances survival in a T cell-dependent manner, reinvigorates T cells, and outperforms individual or sequential therapies in mice. Elevated PD-L1 expression in BRAF-mutant versus BRAF-wild-type glioblastoma supports the rationale for PD-1 inhibition in patients. These findings underscore the potential of targeting glioma plasticity and highlight combination strategies to overcome therapy resistance in BRAFV600E-mutant high-grade glioma.
Keywords: BRAF V600E; BRAF and MEK inhibitor adaptation; Galectin-3; T cell modulation; cell state transitions; high-grade glioma; immune checkpoint inhibition; programmed death-ligand 1.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures






Update of
-
BRAF/MEK Inhibition Induces Cell State Transitions Boosting Immune Checkpoint Sensitivity in BRAFV600E -mutant Glioma.bioRxiv [Preprint]. 2025 Apr 1:2023.02.03.526065. doi: 10.1101/2023.02.03.526065. bioRxiv. 2025. Update in: Cell Rep Med. 2025 Jun 17;6(6):102183. doi: 10.1016/j.xcrm.2025.102183. PMID: 39416185 Free PMC article. Updated. Preprint.
References
-
- Ostrom Q.T., de Blank P.M., Kruchko C., Petersen C.M., Liao P., Finlay J.L., Stearns D.S., Wolff J.E., Wolinsky Y., Letterio J.J., Barnholtz-Sloan J.S. Alex's Lemonade Stand Foundation Infant and Childhood Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2007-2011. Neuro Oncol. 2015;16:x1–x36. doi: 10.1093/neuonc/nou327. Suppl 10. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

