Identification of an Immune-Related Gene Signature for Prognostic Prediction in Glioblastoma: Insights from Integrated Bulk and Single-Cell RNA Sequencing
- PMID: 40507280
- PMCID: PMC12153616
- DOI: 10.3390/cancers17111799
Identification of an Immune-Related Gene Signature for Prognostic Prediction in Glioblastoma: Insights from Integrated Bulk and Single-Cell RNA Sequencing
Abstract
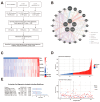
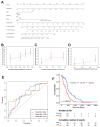
Background: Glioblastoma is a highly malignant brain tumor with limited treatment options and a poor prognosis, largely driven by its complex immune microenvironment. This study aimed to identify and characterize an immune-related gene signature associated with prognosis and immune regulation in glioblastoma. Methods: We performed integrative analyses using bulk and single-cell RNA sequencing data to identify prognostically significant immune-related genes. A five-gene signature (THEMIS2, SIGLEC9, CSTA, LILRB3, and MS4A6A) was derived and its expression patterns were analyzed in association with immune cell infiltration and macrophage subtypes. Functional enrichment and pathway analyses were conducted, followed by drug sensitivity profiling to explore potential therapeutic implications. Results: The five-gene signature was significantly associated with worse survival outcomes and increased immune cell infiltration. Functional analyses revealed involvement in key immune pathways, including antigen presentation, cytokine signaling, and immune cell activation. Single-cell RNA sequencing demonstrated high expression of the signature in tumor-associated macrophages, particularly immune-suppressive and proliferation-associated subtypes. The high expression in proliferation TAMs suggests a role in promoting tumor angiogenesis and growth. Drug sensitivity analysis revealed distinct vulnerabilities between high- and low-risk groups based on signature expression. Conclusions: This Macrophage-Associated Prognostic Signature (MAPS) provides new insights into glioblastoma immunobiology and identifies potential biomarkers and therapeutic targets. It may serve as a valuable tool to guide personalized immunotherapy-based strategies for glioblastoma patients.
Keywords: angiogenesis; glioblastoma; immune signature; macrophages; prognosis; proliferation TAM; single-cell RNA-seq; tumor microenvironment.
Conflict of interest statement
The authors declare no conflicts of interest or relationships that could have influenced the findings or conclusions of this study.
Figures






References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

