Development of a Novel Biomarker Platform for Profiling Key Protein-Protein Interactions to Predict the Efficacy of BH3-Mimetic Drugs
- PMID: 40507334
- PMCID: PMC12153628
- DOI: 10.3390/cancers17111852
Development of a Novel Biomarker Platform for Profiling Key Protein-Protein Interactions to Predict the Efficacy of BH3-Mimetic Drugs
Abstract
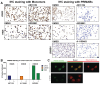
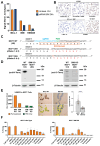
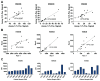
One of the hallmarks of cancer cells is their failure to respond to the cellular mechanism of apoptosis. The B-cell lymphoma 2 (BCL-2) family of proteins regulate apoptosis. Their ability to do so can be measured using several methods that in turn anticipate the fate of the cancer cell in response to apoptosis-inducing treatment. These assays ultimately identify the readiness of the cancer cell to undergo apoptosis, which is referred to as the mitochondrial priming state. These metrics, however, have been challenging to implement in the clinic. Methods: Here, we describe a unique method that relies on a panel of novel conformation-specific antibodies (termed PRIMAB) that can directly measure the mitochondrial priming state. These reagents are highly specific for complexes of their corresponding pro-survival protein interactions with the pro-apoptotic protein BIM. These BIM-containing heterodimeric complexes have long been established as hallmarks of primed cancer cells. Results: Using clinically amenable assay formats, PRIMABs were shown to detect the presence of these anti-apoptotic-pro-apoptotic complexes and their disruption by BH3-mimetic drugs. Moreover, PRIMABs were able to detect a shift in priming status following BH3-mimetic treatment, a factor associated with resistance to these drugs. In a panel of AML patient samples, we report a wide range of priming levels for each PRIMAB complex, demonstrating the potential for heterogeneity in responses. We also show that PRIMABs could be predictive of outcomes for AML patients following cytarabine-based treatment. Conclusions: PRIMABs provide novel and useful tools for cancer research and for clinical implementation as reagents providing predictive tests for treatment response.
Keywords: AML; BH3 domain; BH3 mimetics; PRIMAB; apoptosis; flow cytometry; mitochondrial priming.
Conflict of interest statement
A.J.K. is an employee of Ampersand Biomedicines. F.R. and M.S. are employees of Eutropics. B.L. receives research funding from Beigene for BCL2 inhibitor study. J.A.W. has a grant from the Department of Defense Neurofibromatosis Research Program (NFRP) with iNFixion Bioscience. K.R.K. is a consultant at Amgen, AstraZeneca, Denovo Biopharma, Sanofi-Aventis, Takeda, and Verastem; owns stock or other ownership at Agios and Berkeley Lights; and received speakers’ bureau participation for Bayer, Celgene, Epizyme, Janssen, Karyopharm, Lilly, Novartis, and Pharmacyclics. M.H.C. is an employee of and owns stock in Eutropics. All other authors have no conflicts.
Figures






References
Grants and funding
LinkOut - more resources
Full Text Sources

