Substrate Activation Efficiency in Active Sites of Hydrolases Determined by QM/MM Molecular Dynamics and Neural Networks
- PMID: 40507908
- PMCID: PMC12154731
- DOI: 10.3390/ijms26115097
Substrate Activation Efficiency in Active Sites of Hydrolases Determined by QM/MM Molecular Dynamics and Neural Networks
Abstract
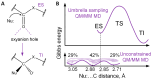
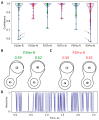
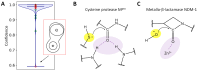
The active sites of enzymes are able to activate substrates and perform chemical reactions that cannot occur in solutions. We focus on the hydrolysis reactions catalyzed by enzymes and initiated by the nucleophilic attack of the substrate's carbonyl carbon atom. From an electronic structure standpoint, substrate activation can be characterized in terms of the Laplacian of the electron density. This is a simple and easily visible imaging technique that allows one to "visualize" the electrophilic site on the carbonyl carbon atom, which occurs only in the activated species. The efficiency of substrate activation by the enzymes can be quantified from the ratio of reactive and nonreactive states derived from the molecular dynamics trajectories executed with quantum mechanics/molecular mechanics potentials. We propose a neural network that assigns the species to reactive and nonreactive ones using the Laplacian of electron density maps. The neural network is trained on the cysteine protease enzyme-substrate complexes, and successfully validated on the zinc-containing hydrolase, thus showing a wide range of applications using the proposed approach.
Keywords: AI; Laplacian of electron density; QM/MM MD; hydrolases; neural network; substrate activation.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

