Vat-Mediated Mucus Penetration Enables Genotoxic Activity of pks+ Escherichia coli
- PMID: 40508162
- PMCID: PMC12155319
- DOI: 10.3390/ijms26115353
Vat-Mediated Mucus Penetration Enables Genotoxic Activity of pks+ Escherichia coli
Abstract
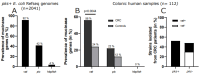
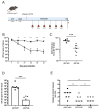
Colibactin toxin-producing Escherichia coli (pks+ E. coli) strains are associated with the occurrence of colorectal cancer in humans. These strains induce DNA damage when in close contact with the cells of the intestinal epithelium. Therefore, maintaining the integrity of the mucus layer that covers the intestinal epithelial mucosa is crucial for counteracting the effects of colibactin. The Vat protein is a mucin protease capable of degrading MUC2 mucus proteins that was previously described in adherent and invasive Escherichia coli strains. Our work shows that the vat gene is found in the genome of all pks+ E. coli strains isolated from patients with colon cancer. In mucus-producing HT29-16E cells, we demonstrated that the Vat protein of E. coli pks+ allows bacteria to penetrate mucus and to reach the epithelial cells. Cells infected with the E. coli pks + vat- strain show a reduction in γ-H2AX staining, a marker of DNA damage. Infection of ApcMin/+ mice with the E. coli pks + vat+ strain or the E. coli pks + vat- mutant revealed that Vat enhances the ability of pks+ E. coli strains to colonize the intestinal mucosa and, in turn, their pro-carcinogenic effects. This study reveals that Vat promotes crossing of the intestinal mucus layer, gut colonization, and the carcinogenicity of pks+ E. coli.
Keywords: Escherichia coli; colibactin; colorectal cancer; mucin; pks.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

