Genome-Wide Identification and Analysis of GATA Gene Family in Dendrobium officinale Under Methyl Jasmonate and Salt Stress
- PMID: 40508251
- PMCID: PMC12157914
- DOI: 10.3390/plants14111576
Genome-Wide Identification and Analysis of GATA Gene Family in Dendrobium officinale Under Methyl Jasmonate and Salt Stress
Abstract
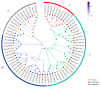
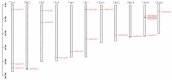
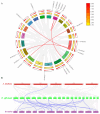
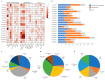
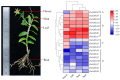
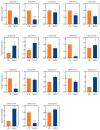
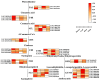
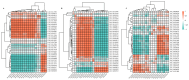
Dendrobium officinale, which was rich in bioactive compounds such as polysaccharides, alkaloids, amino acids, and flavonoids, had significant medicinal value and ability to resist stresses. Studies had demonstrated that GATA genes were one of the crucial regulators in controlling plant development and growth and stress response. Genome-wide identification and characterization of the 18 DoGATA genes were displayed. According to phylogenetic relationships, the DoGATA family genes were divided into 4 groups and the conserved motifs of DoGATA1-DoGATA18 within the same group were similar. All DoGATA genes were localized in the nucleus and randomly mapped on 10 chromosomes. The GATA genes in D. officinale experienced one pair of tandem duplication and 4 pairs of segment duplications to expand the family genes. Additionally, we found that the 2000 bp upstream promoter region of the DoGATA genes harbored 23 types of cis-acting elements that were categorized into plant growth and development, phytohormone responsiveness, and stress responsiveness. DoGATA1-DoGATA18 were diversely expressed across different tissues (root, leaf, stem, flower), exposed to salt stress, and following MeJA treatment. Co-expression analysis between DoGATA and enzyme-encoding genes involved in the biosynthesis of flavone showed that DoCHI (LOC110104562) and DoGTMT (LOC110098370) may be potential downstream targets of DoGATA16 to regulate flavonoid biosynthesis to adapt to salt stress. Furthermore, we confirmed that DoGATA16 may act as a key member to resist stress. The collective findings of this study shed light on the function of GATA genes and molecular breeding of D. officinale.
Keywords: Dendrobium officinale; GATA; expression analysis; flavonoid; stress.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Comparative analysis of B-BOX genes and their expression pattern analysis under various treatments in Dendrobium officinale.BMC Plant Biol. 2019 Jun 10;19(1):245. doi: 10.1186/s12870-019-1851-6. BMC Plant Biol. 2019. PMID: 31182022 Free PMC article.
-
Genome-wide identification and expression analysis of growth-regulating factors in Dendrobium officinale and Dendrobium chrysotoxum.PeerJ. 2023 Dec 15;11:e16644. doi: 10.7717/peerj.16644. eCollection 2023. PeerJ. 2023. PMID: 38111654 Free PMC article.
-
Comparative transcriptomic analysis reveal the regulation mechanism underlying MeJA-induced accumulation of alkaloids in Dendrobium officinale.J Plant Res. 2019 May;132(3):419-429. doi: 10.1007/s10265-019-01099-6. Epub 2019 Mar 22. J Plant Res. 2019. PMID: 30903398
-
Genome-Wide Identification and Functional Characterization of the Dof Family in Dendrobium officinale.Int J Mol Sci. 2025 Mar 16;26(6):2671. doi: 10.3390/ijms26062671. Int J Mol Sci. 2025. PMID: 40141313 Free PMC article.
-
Research Advances in Multi-Omics on the Traditional Chinese Herb Dendrobium officinale.Front Plant Sci. 2022 Jan 11;12:808228. doi: 10.3389/fpls.2021.808228. eCollection 2021. Front Plant Sci. 2022. PMID: 35087561 Free PMC article. Review.
References
-
- Sun J., Qiu C., Ding Y., Wang Y., Sun L., Fan K., Gai Z., Dong G., Wang J., Li X., et al. Fulvic acid ameliorates drought stress-induced damage in tea plants by regulating the ascorbate metabolism and flavonoids biosynthesis. BMC Genom. 2020;21:411. doi: 10.1186/s12864-020-06815-4. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

