Comparative Analysis of Proteomic Characteristics in Seminal Plasma Between Horses and Donkeys
- PMID: 40508997
- PMCID: PMC12153549
- DOI: 10.3390/ani15111532
Comparative Analysis of Proteomic Characteristics in Seminal Plasma Between Horses and Donkeys
Abstract
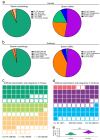
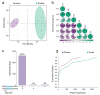
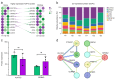
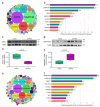
Horses and donkeys, as integral members of the equine family, exhibit distinct reproductive capabilities and characteristics. Seminal plasma, the fluid component of semen, contains a variety of proteins that play critical roles in sperm function and fertility. This study aimed to systematically compare the protein profiles in the seminal plasma of horses and donkeys, thereby elucidating the molecular differences between these two species. The study utilized 4D-DIA proteomics technology to analyze seminal plasma from horses and donkeys and further validated key proteins through Western blot. Our findings revealed significant variations in seminal plasma protein composition between horses and donkeys. We identified 2380 and 2385 proteins in the seminal plasma of horses and donkeys. Among these proteins, 59 are solely present in the seminal plasma of horses, and 64 uniquely exsit in that of donkeys, respectively. These insights enhance our understanding of the biological mechanisms underlying the reproductive distinctions between these equine species. Moreover, the identified species specific proteins may be essential for thier sperm quality and function, which holds practical value for breeding programs and investigations.
Keywords: 4D-DIA; donkey; equid; horse; pregnancy rate; proteomic profile; seminal plasma.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Ren X., Liu Y., Zhao Y., Li B., Bai D., Bou G., Zhang X., Du M., Wang X., Bou T., et al. Analysis of the Whole-Genome Sequences from an Equus Parent-Offspring Trio Provides Insight into the Genomic Incompatibilities in the Hybrid Mule. Genes. 2022;13:2188. doi: 10.3390/genes13122188. - DOI - PMC - PubMed
Grants and funding
- BR220402/the Basic scientific research operating expenses project of universities directly under the Inner Mongolia Autonomous Region
- NJZY23117/the science and technology research projects of universities In Inner Mongolia Autonomous Region
- DC2300001263/the 2022 annual high-level talent research support fund project for local public institutions in Inner Mongolia Autonomous Region
- NDYB2022-42/the research initiation grant program at Inner Mongolia Agricultural University
LinkOut - more resources
Full Text Sources

