Novel Antimicrobials from Computational Modelling and Drug Repositioning: Potential In Silico Strategies to Increase Therapeutic Arsenal Against Antimicrobial Resistance
- PMID: 40509191
- PMCID: PMC12155768
- DOI: 10.3390/molecules30112303
Novel Antimicrobials from Computational Modelling and Drug Repositioning: Potential In Silico Strategies to Increase Therapeutic Arsenal Against Antimicrobial Resistance
Abstract
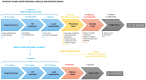
Antimicrobial resistance (AMR) is one of the most significant public health threats today. The need for new antimicrobials against multidrug-resistant infections is growing. The development of computational models capable of predicting new drug-target interactions is an interesting strategy to reposition already known drugs into potential antimicrobials. The objective of this review was to compile the latest advances in the development of computational models capable of identifying drugs already registered by the Food and Drug Administration for other indications with potential capacity to be applied as antimicrobials. We present studies that apply in silico methods such as machine learning, molecular docking, molecular dynamics and deep learning. Some of these studies have in vitro/in vivo results that demonstrate the reliability of this computational methodology in terms of the identification of effective molecules and new targets of interest in the treatment of infections. In addition, we present the methods that are under development and their future prospects in terms of the search for new antimicrobials. We highlight the need to implement these strategies in the research of effective drugs in the treatment of infectious diseases and to continue to improve the available models and approaches to gain an advantage against the rapid emergence of AMR.
Keywords: QSAR models; antimicrobial resistance; computational models; deep learning; drug repositioning; machine learning; mathematical prediction models; molecular docking; molecular dynamics; topological data analysis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Lorente-Torres B., Llano-Verdeja J., Castañera P., Ferrero H.Á., Fernández-Martínez S., Javadimarand F., Mateos L.M., Letek M., Mourenza Á. Innovative Strategies in Drug Repurposing to Tackle Intracellular Bacterial Pathogens. Antibiotics. 2024;13:834. doi: 10.3390/antibiotics13090834. - DOI - PMC - PubMed
-
- Kaur H., Kalia M., Chaudhary N., Singh V., Yadav V.K., Modgil V., Kant V., Mohan B., Bhatia A., Taneja N. Repurposing of FDA approved drugs against uropathogenic Escherichia coli: In silico, in vitro, and in vivo analysis. Microb. Pathog. 2022;169:105665. doi: 10.1016/j.micpath.2022.105665. - DOI - PubMed
-
- Zheng S., Gu Y., Gu Y., Zhao Y., Li L., Wang M., Jiang R., Yu X., Chen T., Li J. Machine learning-enabled virtual screening indicates the anti-tuberculosis activity of aldoxorubicin and quarfloxin with verification by molecular docking, molecular dynamics simulations, and biological evaluations. Brief. Bioinform. 2024;26:696. doi: 10.1093/bib/bbae696. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

