Targeted single cell expression profiling identifies integrators of sleep and metabolic state
- PMID: 40509864
- PMCID: PMC12239624
- DOI: 10.1093/g3journal/jkaf079
Targeted single cell expression profiling identifies integrators of sleep and metabolic state
Abstract
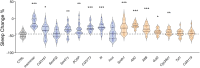
Animals modulate sleep in accordance with their internal and external environments. Metabolic cues are particularly potent regulators of sleep, allowing animals to alter their sleep timing and amount depending on food availability and foraging duration. The fruit fly, Drosophila melanogaster, suppresses sleep in response to acute food deprivation, presumably to forage for food. This process is dependent on a single pair of Lateral Horn Leucokinin (LHLK) neurons, that secrete the neuropeptide Leucokinin. These neurons signal to insulin producing cells and suppress sleep under periods of starvation. The identification of individual neurons that modulate sleep-metabolism interactions provides the opportunity to examine the cellular changes associated with sleep modulation. Here, we use single-cell sequencing of LHLK neurons to examine the transcriptional responses to starvation. We validate that a targeted single-cell sequencing approach selectively isolates RNA from individual LHLK neurons. Single-cell CEL-Seq comparisons of LHLK neurons between fed and 24-h starved flies identified 24 genes that are differentially expressed in accordance with starvation state. In total, 12 upregulated genes and 12 downregulated genes were identified. Gene-ontology analysis showed an enrichment for Attacins, a family of antimicrobial peptides, along with a number of transcripts with diverse roles in regulating cellular function. Targeted knockdown of differentially expressed genes identified multiple genes that function within LHLK neurons to regulate sleep-metabolism interactions. Functionally validated genes include an essential role for the E3 ubiquitin ligase insomniac, the sorbitol dehydrogenase Sodh1, as well as AttacinC and AttacinB in starvation-induced sleep suppression. Taken together, these findings provide a pipeline for identifying novel regulators of sleep-metabolism interactions within individual neurons.
Keywords: feeding; metabolism; single-cell profiling; sleep.
© The Author(s) 2025. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest: The author(s) declare no conflict of interest.
Figures



Update of
-
Targeted single cell expression profiling identifies integrators of sleep and metabolic state.bioRxiv [Preprint]. 2024 Sep 27:2024.09.25.614841. doi: 10.1101/2024.09.25.614841. bioRxiv. 2024. Update in: G3 (Bethesda). 2025 Jul 9;15(7):jkaf079. doi: 10.1093/g3journal/jkaf079. PMID: 39386468 Free PMC article. Updated. Preprint.
References
-
- Blighe K, Rana S, Lewis M. 2018. EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
