Integrated multi-omics analysis and experimental investigation of mitochondrial dynamics-related genes: molecular subtypes, immune landscape, and prognostic implications in lung adenocarcinoma
- PMID: 40510359
- PMCID: PMC12159055
- DOI: 10.3389/fimmu.2025.1585505
Integrated multi-omics analysis and experimental investigation of mitochondrial dynamics-related genes: molecular subtypes, immune landscape, and prognostic implications in lung adenocarcinoma
Abstract
Background: Lung adenocarcinoma (LUAD) is a common and aggressive subtype of lung cancer associated with poor clinical outcomes. The role of mitochondrial dynamics (MD)-related genes in tumor progression and immune regulation remains poorly understood.
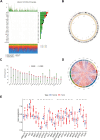
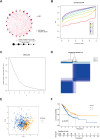
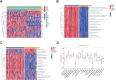
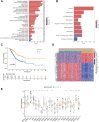
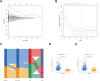
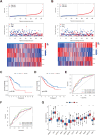
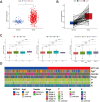
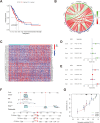
Methods: Data from public databases were integrated, and subtypes were classified based on 23 MD-related genes. A five-gene prognostic model was constructed. Associations between the model and immune infiltration, tumor mutational burden (TMB), tumor stemness, and drug sensitivity were analyzed. The function of the key gene MTCH2 was validated through in vitro experiments.
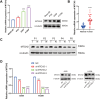
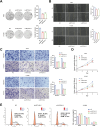
Results: Two distinct MD molecular subtypes were identified, exhibiting significant differences in prognosis and immune characteristics. A corresponding risk score model was established. Patients in the low-risk group showed better prognosis and enhanced immune activity, whereas the high-risk group displayed higher TMB and stemness scores. Drug sensitivity analysis revealed distinct responses to chemotherapeutic agents such as cisplatin and docetaxel between risk groups. Functional assays demonstrated that MTCH2 knockout significantly inhibited LUAD cell proliferation, migration, and invasion, and induced G0/G1 phase arrest, suggesting that MTCH2 may act as a potential adverse prognostic marker.
Conclusion: MD-related genes exhibit strong prognostic and immune subtyping value. The proposed risk model holds clinical potential, and MTCH2 may serve as a promising target for precision therapy in LUAD.
Keywords: LUAD; MTCH2; mitochondrial dynamics; prognostic model; tumor microenvironment.
Copyright © 2025 Wu, Pan, Zeng, Pan, Yu, Lin, Zhang and Jiang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Multi-omics profiling and experimental verification of tertiary lymphoid structure-related genes: molecular subgroups, immune infiltration, and prognostic implications in lung adenocarcinoma.Front Immunol. 2024 Sep 19;15:1453220. doi: 10.3389/fimmu.2024.1453220. eCollection 2024. Front Immunol. 2024. PMID: 39364403 Free PMC article.
-
Integrating multi-omics and machine learning survival frameworks to build a prognostic model based on immune function and cell death patterns in a lung adenocarcinoma cohort.Front Immunol. 2024 Sep 13;15:1460547. doi: 10.3389/fimmu.2024.1460547. eCollection 2024. Front Immunol. 2024. PMID: 39346927 Free PMC article.
-
Multiomics Analysis of Disulfidptosis Patterns and Integrated Machine Learning to Predict Immunotherapy Response in Lung Adenocarcinoma.Curr Med Chem. 2024;31(25):4034-4055. doi: 10.2174/0109298673313281240425050032. Curr Med Chem. 2024. PMID: 38685772
-
Fatty acid metabolism prognostic signature predicts tumor immune microenvironment and immunotherapy, and identifies tumorigenic role of MOGAT2 in lung adenocarcinoma.Front Immunol. 2024 Oct 16;15:1456719. doi: 10.3389/fimmu.2024.1456719. eCollection 2024. Front Immunol. 2024. PMID: 39478862 Free PMC article.
-
Multi-omics characterization and machine learning of lung adenocarcinoma molecular subtypes to guide precise chemotherapy and immunotherapy.Front Immunol. 2024 Nov 28;15:1497300. doi: 10.3389/fimmu.2024.1497300. eCollection 2024. Front Immunol. 2024. PMID: 39669580 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

