Comprehensive bioinformatics analysis and experimental verification identify mitochondrial gene Dgat2 as a novel therapeutic biomarker for myocardial ischemia-reperfusion
- PMID: 40510478
- PMCID: PMC12159077
- DOI: 10.3389/fendo.2025.1539646
Comprehensive bioinformatics analysis and experimental verification identify mitochondrial gene Dgat2 as a novel therapeutic biomarker for myocardial ischemia-reperfusion
Abstract
Background: Ischemic cardiomyopathy is a severe disease marked by high morbidity and mortality, often exacerbated by myocardial ischemia/reperfusion injury (MI/RI). Mitochondrial metabolism plays a critical role in MI/RI progression. This study aimed to identify potential new targets and biomarkers for mitochondria-related genes in MI/RI.
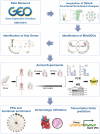
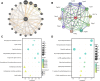
Methods: MI/R microarray data (GSE160516) from the GEO database and a mitochondrial geneset were analyzed. Limma identified differentially expressed genes (DEGs), followed by GSEA, GO, and KEGG pathway enrichment. Mitochondria-related DEGs (MitoDEGs) were pinpointed. Protein-Protein Interaction (PPI) networks and machine learning identified key MitoDEGs. Regulatory networks were constructed using transcription factor (TF) predictions. Immune cell infiltration was assessed with ImmuCelAl, and correlations between MitoDEGs and immune cell levels were examined. Mouse myocardial ischemia-reperfusion models were established to validate pivotal MitoDEGs.
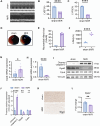
Results: MitoDEGs were enriched in bio-oxidation, immune-inflammation, and oxidative stress pathways. Machine learning identified two hub genes: Dgat2 and Cybb. Dgat2 was significantly elevated in ischemia-reperfusion mouse models, confirmed by RT-PCR and Western blot. Functional enrichment indicated that Dgat2 may be involved in biological oxidation and lipid metabolism. TF prediction suggested PPARG as a regulator of Dgat2 expression. Immune infiltration analysis revealed significant correlations between Dgat2 and immune cells, including CD4_T_cells and NK cells, suggesting a role for immunity in MI/RI.
Conclusions: We found that Dgat2 could be exploited as a novel mitochondria-related gene target and biomarker in myocardial ischemia-reperfusion injury, which is of great clinical significance.
Keywords: Dgat2; bioinformatics analysis; mitochondria; myocardial ischemia/reperfusion injury; oxidative stress.
Copyright © 2025 Li, Zhou, Xue, Yin, Liu, Wu, Zhao, An and Sun.
Conflict of interest statement
The authors state that the study was done without any commercial or financial links that could be seen as a potential conflict of interest.
Figures





Similar articles
-
Exploring the role of mitochondrial metabolism and immune infiltration in myocardial infarction: novel insights from bioinformatics and experimental validation.Front Immunol. 2025 May 27;16:1543584. doi: 10.3389/fimmu.2025.1543584. eCollection 2025. Front Immunol. 2025. PMID: 40496861 Free PMC article.
-
Identification of markers correlating with mitochondrial function in myocardial infarction by bioinformatics.PLoS One. 2024 Dec 30;19(12):e0316463. doi: 10.1371/journal.pone.0316463. eCollection 2024. PLoS One. 2024. PMID: 39775580 Free PMC article.
-
Screening mitochondria-related biomarkers in skin and plasma of atopic dermatitis patients by bioinformatics analysis and machine learning.Front Immunol. 2024 May 7;15:1367602. doi: 10.3389/fimmu.2024.1367602. eCollection 2024. Front Immunol. 2024. PMID: 38774875 Free PMC article.
-
Role of mitochondrial metabolic disorder and immune infiltration in diabetic cardiomyopathy: new insights from bioinformatics analysis.J Transl Med. 2023 Feb 1;21(1):66. doi: 10.1186/s12967-023-03928-8. J Transl Med. 2023. PMID: 36726122 Free PMC article.
-
Role of M6a Methylation in Myocardial Ischemia-Reperfusion Injury and Doxorubicin-Induced Cardiotoxicity.Cardiovasc Toxicol. 2024 Sep;24(9):918-928. doi: 10.1007/s12012-024-09898-7. Epub 2024 Jul 18. Cardiovasc Toxicol. 2024. PMID: 39026038 Review.
References
-
- Jennings RB, Steenbergen C, Jr., Reimer KA. Myocardial ischemia and reperfusion. Monogr Pathol. (1995) 37:47–80. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

