Metabolic reprogramming and computation-aided protein engineering for high-level de novo biosynthesis for 2-phenylethanol in Pichia pastoris
- PMID: 40510534
- PMCID: PMC12159296
- DOI: 10.1016/j.synbio.2025.05.004
Metabolic reprogramming and computation-aided protein engineering for high-level de novo biosynthesis for 2-phenylethanol in Pichia pastoris
Abstract
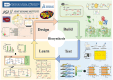
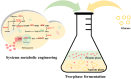
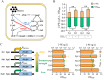
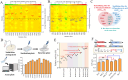
2-Phenylethanol (2-PE), an aromatic compound with a characteristic rose fragrance, is extensively used in the food and cosmetic industries as a flavoring and fragrance agent. Due to limitations in obtaining 2-PE from natural plant sources, microbial cell factories offer a promising alternative for sustainable biosynthesis. In this study, Pichia pastoris was engineered to efficiently synthesize 2-PE. Using computer-assisted predictions of interactions between the key phenylpyruvate decarboxylase KDC2 and its substrates or products, an optimal enzyme variant was rationally designed to boost 2-PE production. Additionally, the shikimic acid pathway was enhanced, and a dynamic regulation promoter was employed to reduce competition from alternative pathways. These strategies significantly increased metabolic flux toward 2-PE production, achieving a titer of 2.81 g/L and 45.8-fold improvement over the non-engineered strain. By integrating controlled carbon feeding and in situ extraction to alleviate acetic acid inhibition and product toxicity, the recombinant strain achieved a final 2-PE titer of 7.10 g/L and a yield of 0.14 g/g glucose, the highest reported microbial production to date. This study highlights the significant potential of P. pastoris as a versatile cell factory for the green biosynthesis of 2-PE and other natural products.
Keywords: 2-Phenylethanol; In situ product removal; Metabolic engineering; Pichia pastoris; Protein engineering.
© 2025 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




Similar articles
-
De novo biosynthesis of 2-phenylethanol in engineered Pichia pastoris.Enzyme Microb Technol. 2020 Feb;133:109459. doi: 10.1016/j.enzmictec.2019.109459. Epub 2019 Nov 1. Enzyme Microb Technol. 2020. PMID: 31874694
-
Multilevel metabolic engineering of Bacillus licheniformis for de novo biosynthesis of 2-phenylethanol.Metab Eng. 2022 Mar;70:43-54. doi: 10.1016/j.ymben.2022.01.007. Epub 2022 Jan 14. Metab Eng. 2022. PMID: 35038552
-
Genetic engineering of an industrial yeast Candida glycerinogenes for efficient production of 2-phenylethanol.Appl Microbiol Biotechnol. 2020 Dec;104(24):10481-10491. doi: 10.1007/s00253-020-10991-4. Epub 2020 Nov 12. Appl Microbiol Biotechnol. 2020. PMID: 33180170
-
[Advances in synthesis of 2-phenylethanol].Sheng Wu Gong Cheng Xue Bao. 2024 Jun 25;40(6):1694-1710. doi: 10.13345/j.cjb.230762. Sheng Wu Gong Cheng Xue Bao. 2024. PMID: 38914486 Review. Chinese.
-
[Advances in biosynthesis of 2-phenylethanol by yeasts].Sheng Wu Gong Cheng Xue Bao. 2016 Sep 25;32(9):1151-1163. doi: 10.13345/j.cjb.150539. Sheng Wu Gong Cheng Xue Bao. 2016. PMID: 29022316 Review. Chinese.
References
-
- Mitri S., Louka N., Rossignol T., Maroun R.G., Koubaa M. Bioproduction of 2-phenylethanol by Yarrowia lipolytica on sugar beet molasses as a low-cost substrate. Fermentation. 2024;10(6):290. doi: 10.3390/fermentation10060290. - DOI
-
- Gao J., Zuo Y., Xiao F., Wang Y., Li D., Xu J., Ye C., Feng L., Jiang L., Liu T. Biosynthesis of catharanthine in engineered Pichia pastoris. Nat Synthesis. 2023;2(3):231–242. doi: 10.1038/s44160-022-00205-2. - DOI
-
- Jin X., Zhang W., Wang Y., Sheng J., Kang Z. Biosynthesis of non-animal chondroitin sulfate from methanol using genetically engineered Pichia pastoris. Green Chem. 2021;23(12):4365–4374. doi: 10.1039/D1GC00260K. - DOI
LinkOut - more resources
Full Text Sources

