Gut Microbiota in Lactose Intolerance: A Mendelian Randomization Study on Microbial Mechanisms and Potential Links to Tumor Inflammatory Microenvironments
- PMID: 40510587
- PMCID: PMC12162160
- DOI: 10.1155/mi/8181816
Gut Microbiota in Lactose Intolerance: A Mendelian Randomization Study on Microbial Mechanisms and Potential Links to Tumor Inflammatory Microenvironments
Abstract
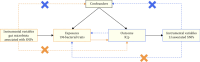
Background: Previous observational studies have suggested an association between the composition of the intestinal microbiome and lactose intolerance (LI). However, the causal direction remains unclear. This study utilized Mendelian randomization (MR) to rigorously evaluate the potential causal link between the gut microbiome and LI. Methods: Genome-wide association studies (GWASs) summary statistics for gut microbiota and LI were sourced from previously published GWAS studies. Multiple methods, such as Simple mode, MR-Egger regression, weighted median, inverse variance-weighted (IVW), and weighted model, were used to determine the causal relationship between gut microbiota and LI. To validate the primary findings from the MR analyses, several sensitivity analyses were conducted. Furthermore, a reverse MR analysis was executed on bacterial taxa previously identified to have a potential causal link with LI risk, aiming to evaluate the possibility of reverse causation. Results: The IVW results revealed that the genus Lachnospiraceae UCG008 (OR = 0.584, 95%CI 0.356-0.958, p=0.0330), genus Eubacterium hallii group (OR = 0.467, 95% CI 0.242-0.899, p=0.023), and genus Ruminococcus gauvreauii group (OR = 0.506, 95% CI 0.2653-0.968, p=0.039) have a protective effect against LI. In contrast, the genus Holdemania (OR = 1.86, 95% CI 1.105-3.131, p=0.0194) displayed a predisposing effect. Sensitivity analyses did not detect any outlier single-nucleotide polymorphisms (SNPs). Further analyses reinforced the association between specific gut microbiota compositions and LI. No evidence suggested reverse causality between LI and the bacterial taxa identified in the reverse MR analysis. Conclusions: From a genetic standpoint, this MR study indicates a causal relationship between variations in gut microbiota composition and LI. This not only underscores the potential of gut microbiota-centric treatments for LI but also provides a foundation for exploring the role of gut microbiota in LI development. Further study of the mechanism of Lachnospiraceae in the treatment of IL is conducive to the discovery of new therapeutic targets for IL.
Keywords: gut microbiota; inflammation; lactose intolerance; mendelian randomization; tumor microenvironment.
Copyright © 2025 Ya Xie et al. Mediators of Inflammation published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Swagerty D. L., Walling A. D., Klein R. M. Lactose Intolerance. American Family Physician . 2002;65(9):1845–1850. - PubMed
-
- Vitellio P., Celano G., Bonfrate L., Gobbetti M., Portincasa P., De Angelis M. Effects of Bifidobacterium Longum and Lactobacillus Rhamnosus on Gut Microbiota in Patients With Lactose Intolerance and Persisting Functional Gastrointestinal Symptoms: A Randomised, Double-Blind, Cross-Over Study. Nutrients . 2019;11(4) doi: 10.3390/nu11040886.886 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

