Exploring current hypervirulent Klebsiella pneumoniae infections: insights into pathogenesis, drug resistance, and vaccine prospects
- PMID: 40510664
- PMCID: PMC12158997
- DOI: 10.3389/fmicb.2025.1604763
Exploring current hypervirulent Klebsiella pneumoniae infections: insights into pathogenesis, drug resistance, and vaccine prospects
Abstract
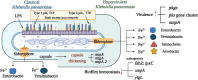
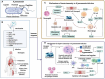
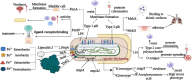
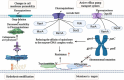
Klebsiella pneumoniae is a significant pathogenic bacterium responsible for a range of infections. The escalating prominence of K. pneumoniae in hospital-acquired infections is a deeply alarming trend that demands immediate attention and rigorous intervention. This article provides an up-to-date review of K. pneumoniae's virulence factors, pathogenesis, and the mechanism driving drug resistance. It also explores the potential for safe and effective vaccine developments, vital for preventing and controlling these diseases. Furthermore, we summarize the epidemiological characteristics of classical and hypervirulent K. pneumoniae infections, providing an overview of drug-resistance K. pneumoniae emergence, transmission, and prevalence.
Keywords: Klebsiella pneumoniae; drug resistance; epidemiology; pathogenesis; vaccine; virulence.
Copyright © 2025 Wang, Yu, Pan, Huang, Lalsiamthara, Ullah, Xu and Lu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Alaubydi M., Al-Saadi D., Ameri L., Fadhil D. (2018). A novel method of preparation attenuated Klebsiella pneumoniae vaccine isolated from respiratory tract infections by using low power laser diodes. J. Mol. Microbiol. Biotechnol. 6 1441–1446.
Publication types
LinkOut - more resources
Full Text Sources

