Expanding the phenotypic and genetic landscape of congenital neutropenia through whole-exome and genome sequencing
- PMID: 40510848
- PMCID: PMC12159251
- DOI: 10.1002/hem3.70150
Expanding the phenotypic and genetic landscape of congenital neutropenia through whole-exome and genome sequencing
Abstract
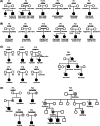
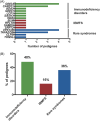
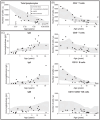
Congenital neutropenia (CN) comprises a heterogeneous group of rare genetic disorders. While some CN cases present only with neutropenia, others present with additional extra-hematological manifestations. The most common cause of CN is variants in ELANE; however, approximately 30 other genes have been implicated. Despite this, the genetic basis remains unknown in roughly 30% of cases. The clinical and genetic heterogeneity of CN makes diagnosis particularly challenging. To address this, we conducted exome or genome sequencing of 60 patients with a suspected diagnosis of CN that remained unresolved following targeted sequencing. A genetic diagnosis was established in 25 patients (42%). Variants were identified in 15 different genes. Half of these cases involved genes traditionally associated with hereditary immunodeficiencies (GINS4, CARD11, ADA2, GINS1, LCP1, SASH3, and WAS). One-third of the cases carried variants in genes linked to syndromic disorders (VPS13B, TAFAZZIN, CLPB, and TONSL), demonstrating variable penetrance of extra-hematological phenotypes. A smaller subset (15%) harbored variants in genes associated with inherited bone marrow failure syndromes (BLM, RPL18, SAMD9, and SRP72), identified incidentally due to atypical presentations. Compared to patients with ELANE-CN, these individuals were diagnosed later, had fewer severe bacterial infections and gingivitis, exhibited less profound neutropenia, lacked monocytosis, and had a granulocytic maturation arrest, often beyond the promyelocytic stage. A shared feature among these cases was a tendency toward reduced lymphocyte subsets, particularly NK cells. This study highlights the significant contribution of exome and genome sequencing in diagnosing CN, given the phenotypic overlap, genetic heterogeneity, and variable penetrance of immunological and extra-hematological features.
© 2025 The Author(s). HemaSphere published by John Wiley & Sons Ltd on behalf of European Hematology Association.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Donadieu J, Beaupain B, Mahlaoui N, Bellanné‐Chantelot C. Epidemiology of congenital neutropenia. Hematol Oncol Clin North Am. 2013;27:1‐17. - PubMed
-
- Donadieu J, Frenz S, Merz L, et al. Chronic neutropenia: how best to assess severity and approach management? Expert Rev Hematol. 2021;14:945‐960. - PubMed
-
- Horwitz M, Benson KF, Person RE, Aprikyan AG, Dale DC. Mutations in ELA2, encoding neutrophil elastase, define a 21‐day biological clock in cyclic haematopoiesis. Nat Genet. 1999;23:433‐436. - PubMed
-
- Dale DC, Person RE, Bolyard AA, et al. Mutations in the gene encoding neutrophil elastase in congenital and cyclic neutropenia. Blood. 2000;96:2317‐2322. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
