Role of Ocrelizumab in modulating gene and microRNA expression in multiple sclerosis
- PMID: 40510879
- PMCID: PMC12159207
- DOI: 10.1016/j.ncrna.2025.05.009
Role of Ocrelizumab in modulating gene and microRNA expression in multiple sclerosis
Abstract
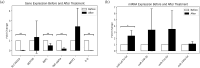
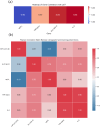
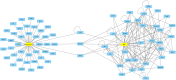
Multiple sclerosis is an autoimmune neurodegenerative disease and one of the most significant challenges in modern neurology, impacting approximately 2.8 million people globally. As a multifactorial condition, susceptibility to MS can result from a combination of genetic and environmental factors. Current treatment strategies aim to prevent acute attacks, slow disease progression, and alleviate symptoms. Ocrelizumab, a monoclonal antibody targeting CD20, has demonstrated efficacy in clinical trials by reducing both disease activity and frequency of relapses. Given the recent approval of this treatment, we investigated whether Ocrelizumab alters the expression of key miRNAs and genes involved in neuroinflammation, such as let-7a-5p, miR-14a-5p, miR-21a-5p, miR-338-3p, IL-1, IL-6, NEAT1, NEFL, NESTIN, SLC16A10 and TNF-alpha, by comparing their expression in patients' blood before and after one year of treatment with Ocrelizumab. Additionally, we explored potential inverse correlations and direct or indirect interactions among the genes and miRNAs that showed significant changes in expression. Lastly, we conducted a pathway analysis to understand the overall effects potentially exerted by the drug. Results revealed a significant decrease in the expression of TNF-alpha, SLC16A10, NEFL and IL-6, and an increase in let-7a-5p expression. There was an inverse correlation between let-7a-5p and the four genes, while the genes positively correlated with each other, suggesting let-7a-5p as a common modulator. These findings indicate that further investigation is needed to determine if the drug directly upregulates let-7a-5p, thereby downregulating the four genes, or if these expression changes are an indication of an overall reduction in inflammation.
Keywords: Inflammation; Interaction network; Multiple sclerosis; Ocrelizumab; Pathway analysis; miRNAs.
© 2025 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

Similar articles
-
Diagnostic Potential of NEAT1, hsa-let-7a-5p, and miR-506-3p in Early-stage Parkinson's Disease.Curr Med Chem. 2024 Oct 31. doi: 10.2174/0109298673336756241016063552. Online ahead of print. Curr Med Chem. 2024. PMID: 39484776
-
Dysregulation of MicroRNAs and Target Genes Networks in Peripheral Blood of Patients With Sporadic Amyotrophic Lateral Sclerosis.Front Mol Neurosci. 2018 Aug 28;11:288. doi: 10.3389/fnmol.2018.00288. eCollection 2018. Front Mol Neurosci. 2018. PMID: 30210287 Free PMC article.
-
Positive regulation of innate immune response by miRNA-let-7a-5p.Front Genet. 2023 Jan 6;13:1025539. doi: 10.3389/fgene.2022.1025539. eCollection 2022. Front Genet. 2023. PMID: 36685889 Free PMC article.
-
Ocrelizumab: A Review in Multiple Sclerosis.Drugs. 2022 Feb;82(3):323-334. doi: 10.1007/s40265-022-01672-9. Epub 2022 Feb 22. Drugs. 2022. PMID: 35192158 Free PMC article. Review.
-
The Epigenetics of Migraine.Int J Mol Sci. 2023 May 23;24(11):9127. doi: 10.3390/ijms24119127. Int J Mol Sci. 2023. PMID: 37298078 Free PMC article. Review.
References
-
- Battaglia M.A., Bezzini D. Estimated prevalence of multiple sclerosis in Italy in 2015. Neurol. Sci. 2017 Mar;38(3):473–479. - PubMed
-
- Ascherio A., Munger K.L. Environmental risk factors for multiple sclerosis. Part II: noninfectious factors. Ann. Neurol. 2007;61:504–513. - PubMed
-
- Rodríguez Murúa S., Farez M.F., Quintana F.J. The immune response in multiple sclerosis. Annu. Rev. Pathol. 2022 Jan;17:121–139. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

