The Centriole Stability Assay: A Method to Investigate Mechanisms Involved in the Maintenance of the Centrosome Structure in Drosophila Cultured Cells
- PMID: 40511405
- PMCID: PMC12152112
- DOI: 10.21769/BioProtoc.5330
The Centriole Stability Assay: A Method to Investigate Mechanisms Involved in the Maintenance of the Centrosome Structure in Drosophila Cultured Cells
Abstract
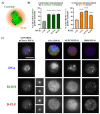
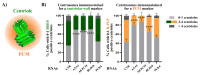
Centrosomes are vital eukaryotic organelles involved in regulating cell adhesion, polarity, mobility, and microtubule (MT) spindle assembly during mitosis. Composed of two centrioles surrounded by the pericentriolar material (PCM), centrosomes serve as the primary microtubule-organizing centers (MTOCs) in proliferating cells. The PCM is crucial for MT nucleation and centriole biogenesis. Centrosome numbers are tightly regulated, typically duplicating once per cell cycle, during the S phase. Deregulation of centrosome components can lead to severe diseases. While traditionally viewed as stable structures, centrosomes can be inactivated or disappear in differentiating cells, such as epithelial cells, muscle cells, neurons, and oocytes. Despite advances in understanding centrosome biogenesis and function, the mechanisms maintaining mature centrosomes or centrioles, as well as the pathways regulating their inactivation or elimination, remain less explored. Studying centrosome maintenance is challenging as it requires the uncoupling of centrosome biogenesis from maintenance. Tools for acute spatial-temporal manipulation are often unavailable, and manipulating multiple components in vivo is complex and time-consuming. This study presents a protocol that decouples centrosome biogenesis from maintenance, allowing the study of critical factors and pathways involved in the maintenance of the integrity of these important cellular structures. Key features • Drosophila cultured cells are resistant to centriole reduplication during S phase arrest, making them a suitable model for studying centrosome integrity without confounding effects from centriole biogenesis.
Keywords: Centrioles; Centrosomes; DMEL-2 cells; Drosophila melanogaster; Maintenance of centrosome integrity.
©Copyright : © 2025 The Authors; This is an open access article under the CC BY-NC license.
Conflict of interest statement
Competing interestsThe authors declare no competing interests.
Figures
Similar articles
-
Ana1/CEP295 is an essential player in the centrosome maintenance program regulated by Polo kinase and the PCM.EMBO Rep. 2024 Jan;25(1):102-127. doi: 10.1038/s44319-023-00020-6. Epub 2024 Jan 10. EMBO Rep. 2024. PMID: 38200359 Free PMC article.
-
The Centrioles, Centrosomes, Basal Bodies, and Cilia of Drosophila melanogaster.Genetics. 2017 May;206(1):33-53. doi: 10.1534/genetics.116.198168. Genetics. 2017. PMID: 28476861 Free PMC article. Review.
-
The mammalian SPD-2 ortholog Cep192 regulates centrosome biogenesis.Curr Biol. 2008 Jan 22;18(2):136-41. doi: 10.1016/j.cub.2007.12.055. Curr Biol. 2008. PMID: 18207742
-
Drosophila Sas-6, Ana2 and Sas-4 self-organise into macromolecular structures that can be used to probe centriole and centrosome assembly.J Cell Sci. 2020 Jun 22;133(12):jcs244574. doi: 10.1242/jcs.244574. J Cell Sci. 2020. PMID: 32409564 Free PMC article.
-
Centrosome Remodelling in Evolution.Cells. 2018 Jul 6;7(7):71. doi: 10.3390/cells7070071. Cells. 2018. PMID: 29986477 Free PMC article. Review.
References
-
- Gönczy P. and Hatzopoulos G. N.(2019). Centriole assembly at a glance. J Cell Sci. 132(4): e228833. https://doi.org/ 10.1242/jcs.228833 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

