Multi-level regulation of hindgut homeostasis by volatile fatty acid administration in dairy goats: linking microbial metabolism to epithelial inflammation and barrier function
- PMID: 40511946
- PMCID: PMC12282142
- DOI: 10.1128/msystems.00116-25
Multi-level regulation of hindgut homeostasis by volatile fatty acid administration in dairy goats: linking microbial metabolism to epithelial inflammation and barrier function
Abstract
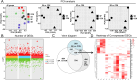
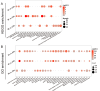
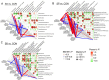
This study provided a comprehensive exploration of the nutritional regulation of volatile fatty acids (VFAs) on hindgut microbial metabolism and epithelial homeostasis in dairy goats. Twenty-four goats were orally administered sodium acetate (SA) at 0.8 g/kg of body weight (BW), propionate (SP) at 0.8 g/kg of BW, butyrate (SB) at 0.5 g/kg of BW, or saline (CON) before morning feeding for 12 days (n = 6/group). Serum and hindgut epithelial tissues were collected to measure antioxidant capacity, inflammatory cytokines, and tight junctions. Cecal contents were collected for 16S rRNA sequencing and metabolome analysis, and colonic epithelial cells were harvested for transcriptome sequencing. The data demonstrated that VFAs positively affected hindgut homeostasis. SB reduced serum malondialdehyde levels (P = 0.042), while SA and SP increased intestinal interleukin-10 concentration compared with CON (P < 0.001). All three VFAs enhanced gut barrier functions by increasing tight junctions compared with CON (P < 0.05). The data revealed distinct bacterial abundances and diversities associated with VFA administration, with notable responders including Rikenellaceae dgA-11 gut group, Christensenellaceae R-7 group, and Bacteroides. Metabolome analysis indicated significant changes in metabolic processes, such as purine, arachidonic acid, tyrosine, and tryptophan metabolism. Transcriptome analysis showed that SA and SP influenced endocrine and digestive functions, metal ion homeostasis, and muscle development in the colonic epithelium, with specific immune response pathways enriched in SB. Correlation analysis suggested interactions between hindgut bacteria and derived metabolites and epithelial homeostasis. In short, our study suggested potential strategies for improving gut health and overall well-being in goats through dietary interventions.
Importance: The volatile fatty acids (VFAs), mainly produced by rumen microbiota, play an important role in ruminal metabolic functions and epithelial health, but their impact on the hindgut has received limited attention. Our study highlighted the significant role of VFAs in hindgut bacterial metabolism and homeostasis, providing novel insights into the role of VFAs in regulating hindgut metabolism and physiological homeostasis beyond the rumen.
Keywords: dairy goats; gut bacteria; gut homeostasis; hindgut; volatile fatty acids.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Ranadheera CS, Evans CA, Baines SK, Balthazar CF, Cruz AG, Esmerino EA, Freitas MQ, Pimentel TC, Wittwer AE, Naumovski N, Graça JS, Sant’Ana AS, Ajlouni S, Vasiljevic T. 2019. Probiotics in goat milk products: delivery capacity and ability to improve sensory attributes. Compr Rev Food Sci Food Saf 18:867–882. doi: 10.1111/1541-4337.12447 - DOI - PubMed
-
- Ranadheera CS, Naumovski N, Ajlouni S. 2018. Non-bovine milk products as emerging probiotic carriers: recent developments and innovations. Curr Opin Food Sci 22:109–114. doi: 10.1016/j.cofs.2018.02.010 - DOI
-
- Nayik GA, Jagdale YD, Gaikwad SA, Devkatte AN, Dar AH, Dezmirean DS, Bobis O, Ranjha M, Ansari MJ, Hemeg HA, Alotaibi SS. 2021. Recent insights into processing approaches and potential health benefits of goat milk and its products: a review. Front Nutr 8:789117. doi: 10.3389/fnut.2021.789117 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 2024YFD1300204, 2023YFD1301705/the National 14th Five-Year Plan Key Research and Development Program
- 31672446/National Natural Science Foundation of China
- KYCX23_3590/the Postgraduate Research & Practice Innovation Program of Jiangsu Province
- the Excellent Doctoral Dissertation Fund of Yangzhou University
- Priority Academic Program Development of Jiangsu Higher Education Institutions
LinkOut - more resources
Full Text Sources

