A novel cuproptosis-associated LncRNA model predicting prognostic and immunotherapy response for glioma
- PMID: 40512434
- PMCID: PMC12165929
- DOI: 10.1007/s12672-025-02912-6
A novel cuproptosis-associated LncRNA model predicting prognostic and immunotherapy response for glioma
Abstract
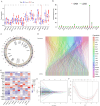
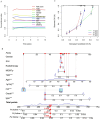
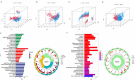
Recent studies have identified cuproptosis as a novel form of regulated cell death (RCD), and long non-coding RNAs (lncRNAs) have been implicated in glioma progression and prognosis. However, the role of cuproptosis-associated lncRNAs in gliomas has not been systematically assessed. In this study, data from the Cancer Genome Atlas (TCGA) and the Chinese Glioma Genome Atlas (CGGA) databases were used, and cuproptosis-related genes were obtained from previous research. Cuproptosis-associated lncRNAs were identified through co-expression network analysis, Cox regression, and Least Absolute Shrinkage and Selection Operator (LASSO). A total of 10 cuproptosis-associated lncRNAs were selected to construct a prognostic prediction model. The high-risk group was associated with poor overall survival (OS) and progression-free survival (PFS). Multivariate Cox regression, Receiver Operating Characteristic (ROC) curve analysis, C-index, and nomogram demonstrated the accuracy of the 10-lncRNA signature in predicting outcomes in glioma patients. Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Gene Set Variation Analysis (GSVA) enrichment analyses revealed a strong association between the signature and immune response pathways. Immune cell infiltration and Single-Sample Gene Set Enrichment Analysis (ssGSEA) further confirmed that the signature is closely linked to immune responses in glioma patients. Further investigation revealed significant differences in tumor immune dysfunction and rejection (TIDE) scores and half-maximal inhibitory concentration (IC50) values for many drugs between low- and high-risk subgroups. This risk signature may serve as a prognostic tool and offer valuable insights into treatment strategies for glioma patients. Additionally, the expression levels of the 10 signature genes were validated by quantitative real-time polymerase chain reaction (qRT-PCR).
Supplementary Information: The online version contains supplementary material available at 10.1007/s12672-025-02912-6.
Keywords: Cuproptosis; Glioma; Immune microenvironment; LncRNA; Prognosis.
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study was approved by the Ethics Committee of Guizhou Provincial People’s Hospital. All procedures performed in this study involving human participants were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki Declaration and its later amendments. Consent to publish: All patients provided consent for publication of anonymized data related to this study. Competing interests: The authors declare no competing interests. Informed consent: Written informed consent was obtained from all patients involved in the study.
Figures





References
-
- Ostrom QT, Price M, Neff C, Cioffi G, Waite KA, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the united States in 2015–2019. Neuro Oncol. 2022;24(Supplement5):v1–95. 10.1093/neuonc/noac202. PMID: 36196752; PMCID: PMC9533228. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
