Aberrant phase separation of two PKA RIβ neurological disorder mutants leads to mechanistically distinct signaling deficits
- PMID: 40512625
- PMCID: PMC12369641
- DOI: 10.1016/j.celrep.2025.115797
Aberrant phase separation of two PKA RIβ neurological disorder mutants leads to mechanistically distinct signaling deficits
Abstract
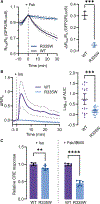
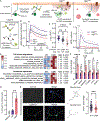
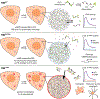
Spatiotemporal regulation of key-node signaling molecules, such as 3',5'-cyclic adenosine monophosphate (cAMP)-dependent protein kinase (PKA), is critical for normal cell physiology and susceptible to dysregulation in disease. Liquid-liquid phase separation (LLPS) is broadly recognized as a fundamental component of signal regulation, yet the connections between physiological and disease-linked biomolecular condensates are not well understood. Here, we show that an understudied, brain-specific PKA regulatory subunit, RIβ, forms biomolecular condensates with distinct features from the ubiquitous isoform, RIα. We demonstrate that two RIβ mutants linked to neurodegenerative (L50R) or neurodevelopmental (R335W) pathologies produce aberrant condensates that trap the PKA catalytic subunit within a gel-like matrix or cAMP-insensitive holoenzyme complex, respectively. RIβL50R condensates, in particular, lead to disrupted spatiotemporal control of PKA signaling and diminished PKA activity, resulting in phenotypic hallmarks of neurodegeneration. Our work highlights the functional importance of biomolecular condensates and the critical link between dysregulated LLPS and neurological disorders.
Keywords: CP: Molecular biology; CP: Neuroscience; biomolecular condensates; cAMP; neurodegeneration; neurodevelopment; protein kinase; signaling compartmentation.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

