From experimental clues to theoretical modeling: Evolution associated with the membrane-takeover at an early stage of life
- PMID: 40512812
- PMCID: PMC12201655
- DOI: 10.1371/journal.pcbi.1012763
From experimental clues to theoretical modeling: Evolution associated with the membrane-takeover at an early stage of life
Abstract
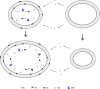
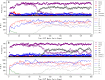
Modern cell membranes are primarily composed of phospholipids, while primitive cell membranes in the beginning of life are believed to have formed from simpler lipids (such as fatty acids) synthesized in the prebiotic environment. An attractive experimental study suggested that the corresponding "membrane-takeover" (as an evolutionary process) is likely to have occurred very early (e.g., in the RNA world) due to some simple physical effects, and might have subsequently triggered some other evolutionary processes. Here, via computer modeling on a system of RNA-based protocells, we convinced the plausibility of such a scenario and elaborated on relevant mechanisms. It is shown that in protocells with a fatty-acid membrane, because of the benefit of phospholipid content (i.e., stabilizing the membrane), a ribozyme favoring the synthesis of phospholipids may emerge; subsequently, due to the reduced membrane permeability on account of the phospholipid content, two other functional RNA species could arise: a ribozyme exploiting more fundamental materials (thus more permeable) for nucleotide synthesis and a species favoring across-membrane transportation. This case exemplifies a combination of experimental and theoretical efforts regarding early evolution, which may shed light on that notoriously complicated problem: the origin of life.
Copyright: © 2025 Ma, Yu. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Fiore M. Prebiotic amphiphiles: the systems chemistry perspective. In: Fiore M, Editor. Prebiotic Chemistry and Life’s Origin. Royal Society of Chemistry. 2022. p. 269–92.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

